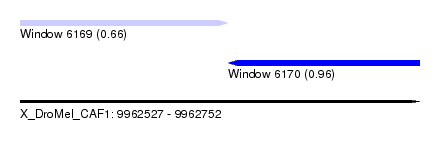
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,962,527 – 9,962,752 |
| Length | 225 |
| Max. P | 0.962540 |

| Location | 9,962,527 – 9,962,644 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9962527 117 + 22224390 GCCUAUUCUUGUGCGCGCUGCUAAUUUACGAGGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGACUC---AGUCUUGGCUUUUAUUCCCCUCAUUCCCCUUUUUACC (((.......((((.(((.((.((((.........)))).)).)))((((........))))...))))..(((((...)---))))..)))............................ ( -24.90) >DroEre_CAF1 25162 98 + 1 GCCUAUUCUUGUGCGCGCUGCUAAUUUACGAGGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGACGC---GGUCUUGGCUUUCAUCCC------------------- (((.......((((.(((.((.((((.........)))).)).)))((((........))))...))))..(((((...)---))))..))).........------------------- ( -24.20) >DroYak_CAF1 25058 108 + 1 GUCUAUUC-UGUGCGCGCUGCUAAUUUACGAGGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGACUCAGUAGUCUUGGCUUUUAUCCC-----------CUUUUACC ........-.((((..(((.(........).)))((((((((....))))))))......)))).((.(..(((((......)))))..))).........-----------........ ( -21.80) >consensus GCCUAUUCUUGUGCGCGCUGCUAAUUUACGAGGCUAAUUUGCUGCGGCAAGUUAUUCAUUGCACUGCACUUGACUGACUC___AGUCUUGGCUUUUAUCCC____________UUUUACC (((...((..((((.(((.((.((((.........)))).)).)))((((........))))...))))..))..(((......)))..)))............................ (-22.52 = -22.30 + -0.22)


| Location | 9,962,644 – 9,962,752 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -25.32 |
| Energy contribution | -27.77 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9962644 108 - 22224390 UUUGAGCUGCCGCU------------UUUUCUUUAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGCAGAGAGACGUGACAGGAAAUGGCCAACAAAACAAAUGGUGGCAUUGGGU .......(((((((------------.(((.(((..((((...((..((((((((..(.(((((..........))))))..))))))))..)).))))))).))).)))))))...... ( -32.60) >DroEre_CAF1 25260 114 - 1 UUUGAGCUGCUGCCUCGCUUGCCCCUCUUUCUUGAUUGUUUUUGCUCUUUUCUGUUGCUGUCUCAUGUGGCAGA--GACGUGACAGGAAAUGGCCAACACAACAAAAGGGGGCAAG---- ...((((....).))).((((((((.((((..((.((((.((.(((.((((((((..(.(((((........))--))))..)))))))).))).)).))))))))))))))))))---- ( -46.50) >DroYak_CAF1 25166 117 - 1 UUUGAGCUGCUGCUUUAGUUGCCCCCUUUUCUUGAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGCAGA--GACGUGACAGGAAAUGGCCAACAAAACAAAAGGGGGCAAG-GGG ...((((....))))...((((((((((((.((..(((((...((..((((((((..(.(((((........))--))))..))))))))..)).))))))).)))))))))))).-... ( -46.30) >consensus UUUGAGCUGCUGCUU___UUGCCCC_UUUUCUUGAUUGUUUUUGUUCUUUUCUGUUGCUGUCUCAUGUGGCAGA__GACGUGACAGGAAAUGGCCAACAAAACAAAAGGGGGCAAG_GG_ .....((....)).....((((((((.(((.(((..((((...((..((((((((..(.(((((........))..))))..))))))))..)).))))))).))).))))))))..... (-25.32 = -27.77 + 2.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:38 2006