
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,957,735 – 9,958,012 |
| Length | 277 |
| Max. P | 0.749026 |

| Location | 9,957,735 – 9,957,854 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -23.13 |
| Energy contribution | -24.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9957735 119 + 22224390 UUUCUUUCUCUCGCGGCUAUUUGCCAUUUGCCAUUUGCCUUUG-GAACUGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCACUCCAUGGUAAACCCAGAAACCCCUAACUUUG ....(((((.....(((.....))).((((((.....((...)-)......((((((((((((...................))))))))))))))))))...)))))............ ( -30.01) >DroSec_CAF1 25766 112 + 1 UCUCUUUCUCUCGCGGCUAUUUGCC-------AUUUGCCUUUG-GAACAGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCACUCCAUGGUAAACCCCGAAACCCCUAACUUUG ..........(((.(((.....)))-------.((((((((((-...))))((((((((((((...................))))))))))))))))))...))).............. ( -28.91) >DroSim_CAF1 6957 115 + 1 ----UUUCUCUCGCGGCUAUUUGCCAUUUGCCAUUUGCCUUUG-GAACAGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCACUCCAUGGUAAACCCCGAAACCCCUAACUUUG ----......(((.((...((((((.(((((((........))-)..))))((((((((((((...................)))))))))))))))))).))))).............. ( -31.31) >DroEre_CAF1 20402 110 + 1 GCUCAUUC--UCUCAGCUAUUUGCC-------AUUUGCCUUUGGGAACAGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCAGUCCAUGGUAAAGCCCGAAACCCCUAACU-UG (((.....--....)))..((((..-------.((((((((((....))))(((((.((((((...................)))))).)))))))))))...))))..........-.. ( -25.31) >DroYak_CAF1 20152 102 + 1 ----------UUUCAGCUAUUUGCC-------AUUUGCCUUUG-GAACAGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCACUCCACGGUAAACCCCGAAACCCCCAACUUUG ----------.....((.....)).-------........(((-(.......(((((((((((...................)))))))))))(((......)))......))))..... ( -24.01) >consensus __UCUUUCUCUCGCGGCUAUUUGCC_______AUUUGCCUUUG_GAACAGAAUGGAGUGGAGCUCCAUGUGUCAUUUUUAUCGCUUCACUCCAUGGUAAACCCCGAAACCCCUAACUUUG ..............(((.....)))........((((((((((....))))((((((((((((...................)))))))))))))))))).................... (-23.13 = -24.13 + 1.00)

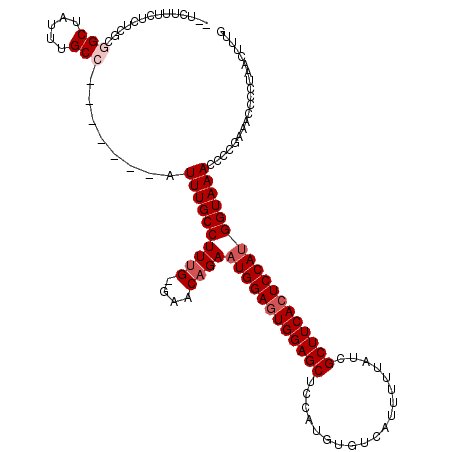
| Location | 9,957,854 – 9,957,972 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -23.94 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9957854 118 + 22224390 CA--ACCACUCCACCAUGCUCUGCUGGCAUCUCGAAUCCGGAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAG .(--((((.......((((....((((.(((((.......)))))))))((((((......))))))))))......(((((((.(......).))).)))).)))))............ ( -31.20) >DroSec_CAF1 25878 120 + 1 CAGCACCACUCCACCCUGCCCUGCUGGCAUCUCGAAUCCGGAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCUGUCUGGCUUGGUUUUUAUGACUCAG ((((.(((((.(((..((((.....)))).......(((((....)))))))).....))))).))))(((((((..((((((((.(((.....))).))).))))))))))))...... ( -39.30) >DroSim_CAF1 7072 120 + 1 CAGCACCACUCCACCCUGCCCUGCUGGCAUCUCGAAUCCGGAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUGCACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAA ((((.(((((.(((..((((.....)))).......(((((....)))))))).....))))).))))(((((((..(((((((((......))))).)))).....)))))))...... ( -44.10) >DroEre_CAF1 20512 106 + 1 C---ACCACUC---CCCGCCCCCUUGGCAUCCCGAAUUC--------GGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGCUUUUUAUGACUCAG .---.......---..((((..(((((((..(((....)--------))..)))..)))).))))...(((((((..(((((((.(......).))).)))).....)))))))...... ( -28.90) >DroYak_CAF1 20254 106 + 1 C---ACCACUC---CCGGCUCUCUUGGCAUCCCGAAUCC--------GGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAG .---.......---.((((((.(((((((..(((....)--------))..)))..)))).)).))))(((((((..((((((((.((.......)).))).))))))))))))...... ( -28.80) >consensus CA__ACCACUCCACCCUGCCCUGCUGGCAUCUCGAAUCCGGAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAG .....(((((.......(((.....))).....(..(((((....)))))...)....))))).....(((((((..(((((((.(......).))).)))).....)))))))...... (-23.94 = -23.94 + -0.00)


| Location | 9,957,892 – 9,958,012 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -34.42 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9957892 120 + 22224390 GAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAGAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG .......(((....)))..(((((((((((......)))...((((.((((((((((..(((((((((.....))).)))....)))....)))))))))).))))..)).)).)))).. ( -36.50) >DroSec_CAF1 25918 120 + 1 GAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCUGUCUGGCUUGGUUUUUAUGACUCAGAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG .......(((....)))..((((((((((((((((..((((((((.(((.....))).))).))))))))))))((((((.((((.....))))....))))))...))).)).)))).. ( -34.90) >DroSim_CAF1 7112 120 + 1 GAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUGCACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAAAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG .......(((....)))..((((((((((((((((..(((((((((......))))).)))).....)))))))((((((.((((.....))))....))))))...))).)).)))).. ( -40.00) >DroEre_CAF1 20545 113 + 1 -------GGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGCUUUUUAUGACUCAGAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG -------(((....)))..(((((((((((......)))...((((.((((((((((..(((..((((((........)))))))))....)))))))))).))))..)).)).)))).. ( -38.00) >DroYak_CAF1 20287 113 + 1 -------GGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAGAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG -------(((....)))..(((((((((((......)))...((((.((((((((((..(((((((((.....))).)))....)))....)))))))))).))))..)).)).)))).. ( -36.20) >consensus GAGAUCCGGAGUGCUCCAAGUGGCGCUGCAUAAAAUUGCCAACUACACAAUCGCAGUCUGGCUUGGUUUUUAUGACUCAGAAGUGCUUUACGCUGUGAUUGAGUAGACAGCCGACCGCAG .......(((....)))..(((((((((((......)))...((((.((((((((((..(((((((((.....))).)))....)))....)))))))))).))))..)).)).)))).. (-34.42 = -34.50 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:27 2006