
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,956,085 – 9,956,193 |
| Length | 108 |
| Max. P | 0.868750 |

| Location | 9,956,085 – 9,956,193 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -16.44 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.02 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
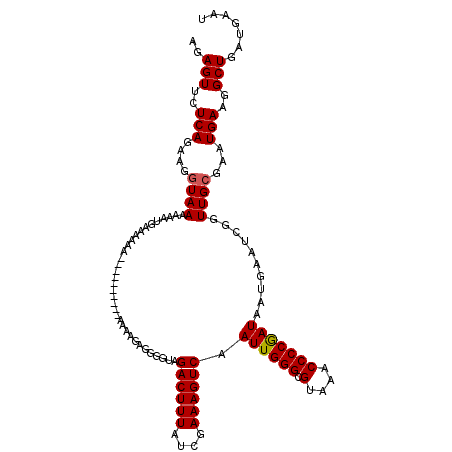
>X_DroMel_CAF1 9956085 108 + 22224390 ------UAGCCUUCAUUCGCAACCGAUUCAUUAUUGGGGUUUACGCCCAAUUGACUUUCGAUAAAGUCUACCCCCCUUCUU------UUUUUUUUUCAUUUUUUACCUUCUGAGAACUCU ------..((........))....(((((...(((((((....).)))))).((((((....)))))).............------..........................))).)). ( -12.80) >DroSec_CAF1 24176 110 + 1 AUUCAUCAGCCUUCAUUCGCAACCGAUUCAUUAUCGGGGUUUACGCCCAAUUGACUUUCGAUAAAGUCUACCCCUCUUUU----------UUUUUUCAUUUUUUACCUUCUGAGAACUCU .(((.((((...............(((.....)))(((((..((.....((((.....))))...))..)))))......----------...................))))))).... ( -16.70) >DroSim_CAF1 5323 120 + 1 AUUCAUCAGCCUUCAUUCGCAACCGAUUCAUUAUCGGGGUUUACGCCCAAUUGACUUUCGAUAAAGUCUACCCCUCUUUUUUUUCAUUUUUUUUUUCAUUUUUUACCUUCUGAGAACUCU .(((.((((...............(((.....)))(((((..((.....((((.....))))...))..)))))...................................))))))).... ( -16.70) >DroEre_CAF1 18773 108 + 1 AUUCAUCAGCCUUCAAUCCCAACCGAUUCAAUAUCGGGGGCUACGCCCAAUUGACUUUCGAUAAAGUCU--UCUGCCCCU----------CUUUUUUAUUUUUUACCUUCUGAGAACUUU .(((.((((.............(((((.....)))))((((...))))....((((((....)))))).--.........----------...................))))))).... ( -20.30) >DroYak_CAF1 18522 108 + 1 AUUCAUCAGCCUUCAUUCGCAACCGAUUCAUUAUCGGGGUUUACGCCCAAUUGACUUUCGAUAAAGUCC--CCCUCUUGU----------UUUAUUUUUUUUUUACCUUCUGAGAACUUU .(((.((((..............((((.....))))((((....))))....((((((....)))))).--.........----------...................))))))).... ( -15.70) >consensus AUUCAUCAGCCUUCAUUCGCAACCGAUUCAUUAUCGGGGUUUACGCCCAAUUGACUUUCGAUAAAGUCUACCCCUCUUCU__________UUUUUUCAUUUUUUACCUUCUGAGAACUCU .....((((..............((((.....))))(((......)))....((((((....)))))).........................................))))....... (-13.14 = -13.02 + -0.12)



| Location | 9,956,085 – 9,956,193 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9956085 108 - 22224390 AGAGUUCUCAGAAGGUAAAAAAUGAAAAAAAAA------AAGAAGGGGGGUAGACUUUAUCGAAAGUCAAUUGGGCGUAAACCCCAAUAAUGAAUCGGUUGCGAAUGAAGGCUA------ ..(((..(((....((((....(((........------.............((((((....)))))).((((((.(....)))))))......))).))))...)))..))).------ ( -21.70) >DroSec_CAF1 24176 110 - 1 AGAGUUCUCAGAAGGUAAAAAAUGAAAAAA----------AAAAGAGGGGUAGACUUUAUCGAAAGUCAAUUGGGCGUAAACCCCGAUAAUGAAUCGGUUGCGAAUGAAGGCUGAUGAAU ..(((..(((....((((....(((.....----------............((((((....)))))).((((((.(....)))))))......))).))))...)))..)))....... ( -21.40) >DroSim_CAF1 5323 120 - 1 AGAGUUCUCAGAAGGUAAAAAAUGAAAAAAAAAAUGAAAAAAAAGAGGGGUAGACUUUAUCGAAAGUCAAUUGGGCGUAAACCCCGAUAAUGAAUCGGUUGCGAAUGAAGGCUGAUGAAU ..(((..(((....((((....(((...........................((((((....)))))).((((((.(....)))))))......))).))))...)))..)))....... ( -21.40) >DroEre_CAF1 18773 108 - 1 AAAGUUCUCAGAAGGUAAAAAAUAAAAAAG----------AGGGGCAGA--AGACUUUAUCGAAAGUCAAUUGGGCGUAGCCCCCGAUAUUGAAUCGGUUGGGAUUGAAGGCUGAUGAAU ...((((((((..................(----------.(((((...--.((((((....)))))).((.....)).))))))....((.((((......)))).))..)))).)))) ( -22.40) >DroYak_CAF1 18522 108 - 1 AAAGUUCUCAGAAGGUAAAAAAAAAAUAAA----------ACAAGAGGG--GGACUUUAUCGAAAGUCAAUUGGGCGUAAACCCCGAUAAUGAAUCGGUUGCGAAUGAAGGCUGAUGAAU ..(((..(((....((((............----------....((((.--...)))).((((...(((((((((.(....)))))))..))).))))))))...)))..)))....... ( -22.70) >consensus AGAGUUCUCAGAAGGUAAAAAAUGAAAAAA__________AAAAGAGGGGUAGACUUUAUCGAAAGUCAAUUGGGCGUAAACCCCGAUAAUGAAUCGGUUGCGAAUGAAGGCUGAUGAAU ..(((..(((....((((..................................((((((....)))))).((((((.(....)))))))..........))))...)))..)))....... (-18.86 = -18.90 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:20 2006