
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,955,599 – 9,955,790 |
| Length | 191 |
| Max. P | 0.990711 |

| Location | 9,955,599 – 9,955,719 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955599 120 - 22224390 GUCGGCCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAAGCCAAAUGAAAAUCUCGCUUGCCAAUUUGUCGUCUCCCUU ...(((.(((((..((((..........((.((((((((((((............))))))))))))))..........))))..((....)).))))).)))................. ( -31.25) >DroSec_CAF1 18131 119 - 1 GUCG-UCGGCGAAAGGUUAACUUUAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAAGUCAAAUGAAAAUCUCUCUUGGCAAUUUGUCGUCUGCCUU .(((-....)))(((((..((..((((.((.((((((((((((............))))))))))))))(((((..(((..((....))...)))...)))))..))))..))..))))) ( -33.20) >DroSim_CAF1 4824 119 - 1 GUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAAGCCAAAUGAAAAUCUCUCUUGGCAAUUUGUCGUCUGCCUU ((.(-.(((((((.(((.....(....)((.((((((((((((............)))))))))))))))))........(((((..((.....))..)))))..))))))).).))... ( -33.30) >DroEre_CAF1 18274 113 - 1 GUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUG------AAUGGCAAUUGAGGCGCCAAACAGAAGCCAAAUGAAAAUCUCUCUUGGCAAUUUGUCGUCUCCCUU ...(-.(((((((.(((.....(....)((.((((((((((((.....------.)))))))))))))))))........(((((..((.....))..)))))..))))))).)...... ( -33.90) >DroYak_CAF1 18024 113 - 1 GUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUG------AAUGGCAAUUGAGGCGCCAAACAGAAGCCAAAUGAAAAUCUCUCUUGGCAAUUUGUCGUCUCCCUU ...(-.(((((((.(((.....(....)((.((((((((((((.....------.)))))))))))))))))........(((((..((.....))..)))))..))))))).)...... ( -33.90) >consensus GUCG_UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAAGCCAAAUGAAAAUCUCUCUUGGCAAUUUGUCGUCUCCCUU .......(((((.(((((..........((.((((((((((((............))))))))))))))...........(((((..((.....))..)))))))))).)))))...... (-29.60 = -29.68 + 0.08)

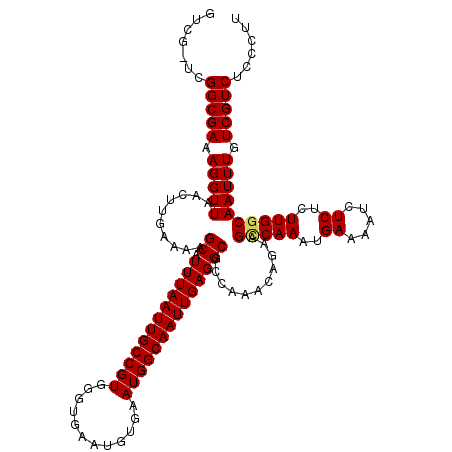
| Location | 9,955,639 – 9,955,751 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955639 112 - 22224390 AUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCGGCCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAA ........(((((((.......)))))))(((((.(((...(....).............((.((((((((((((............)))))))))))))))))..))))). ( -29.70) >DroSim_CAF1 4864 111 - 1 AUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUGAAUGUGAAUGGCAAUUGAGGCGCCAAACAGAA ........(((((((.......)))))))(((((..-..(((..(((.....))).....((.((((((((((((............)))))))))))))))))..))))). ( -29.20) >DroEre_CAF1 18314 105 - 1 AUCGAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUG------AAUGGCAAUUGAGGCGCCAAACAGAA ..(((..((((((((.......))))))))...)))-..(((..(((.....))).....((.((((((((((((.....------.)))))))))))))))))........ ( -30.90) >DroYak_CAF1 18064 105 - 1 AUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUG------AAUGGCAAUUGAGGCGCCAAACAGAA ........(((((((.......)))))))(((((..-..(((..(((.....))).....((.((((((((((((.....------.)))))))))))))))))..))))). ( -30.50) >consensus AUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG_UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGCCGUGGGUG______AAUGGCAAUUGAGGCGCCAAACAGAA ........(((((((.......)))))))(((((.....(((..(((.....))).....((.((((((((((((............)))))))))))))))))..))))). (-28.50 = -28.50 + 0.00)

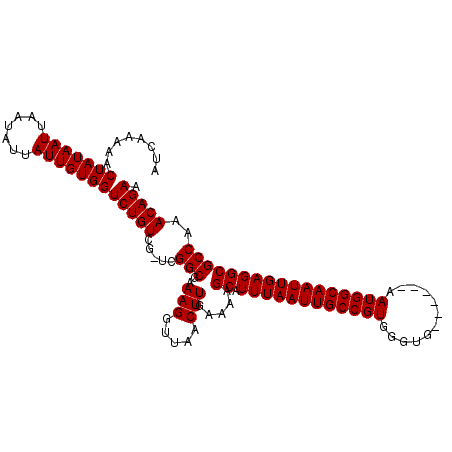

| Location | 9,955,679 – 9,955,790 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.56 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -23.46 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955679 111 - 22224390 CAGGCUGCUUCUUUAAUUGAAUGCCCAUUCAAGU-CGAGCAUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCGGCCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGC .............(((((((((((...(((((((-..(.(.......((((((((.......)))))))).((((....))))...).)..)))))))..))))))))))). ( -27.00) >DroSim_CAF1 4904 111 - 1 CAGGCUGCUUCUUUAAUUGAAUGCCCAUUCAAGUCCGAGCAUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGC .............(((((((((((...(((((((((((.(..((...((((((((.......)))))))).))..)-))))(....)....)))))))..))))))))))). ( -27.50) >DroEre_CAF1 18348 110 - 1 CAGGCUUCUUCUUUAAUUGAAUGCCCAUUCAAG-CCGAGCAUCGAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGC .............(((((((((((...((((((-((((....(((..((((((((.......))))))))...)))-))))(....).....))))))..))))))))))). ( -28.10) >DroYak_CAF1 18098 110 - 1 CAGGCUUCUUCUUUAAUUGAAUGCCCAUUCAAC-CCGAGCAUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG-UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGC .............(((((((((((...(((((.-((((.(..((...((((((((.......)))))))).))..)-))))(....)......)))))..))))))))))). ( -23.60) >consensus CAGGCUGCUUCUUUAAUUGAAUGCCCAUUCAAG_CCGAGCAUCAAAAACUAUAAUUAAUAUUAUUGUGGUCUGUCG_UCGGCGAAAGGUUAACUUGAAAAGCAUUUAAUUGC .............(((((((((((...((((((.((((.(..((...((((((((.......)))))))).))..).))))(....).....))))))..))))))))))). (-23.46 = -24.02 + 0.56)

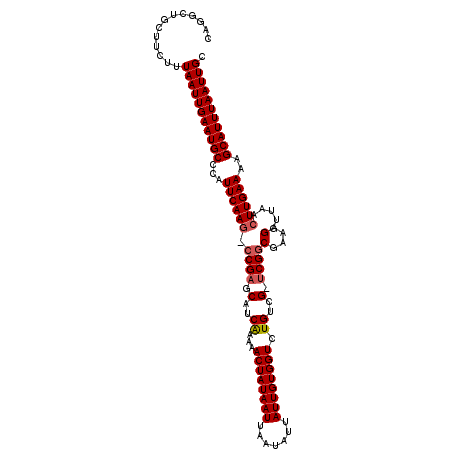

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:19 2006