
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,954,943 – 9,955,149 |
| Length | 206 |
| Max. P | 0.999785 |

| Location | 9,954,943 – 9,955,053 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9954943 110 - 22224390 UAUAAU----------UCGUGUGCAUAUGUAACACUUAUAUAAUGCACUUGGUUUCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA ......----------.((.(((((((((((.....))))).)))))).))((((((((.....(((((((.....((((....(((......)))..))))..))))))))))))))). ( -23.70) >DroSec_CAF1 17482 110 - 1 UGUAAU----------UGGUGUGCAUAUGCAACACUUAUAUAAUGCACUUGGUUUCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA ((((..----------.(((((((....)).)))))..)))).........((((((((.....(((((((.....((((....(((......)))..))))..))))))))))))))). ( -21.60) >DroSim_CAF1 4179 110 - 1 UGUAAU----------UCGUGUGCAUAUGCAACACUUAUAUAAUGCACUUGGUUUCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA ......----------.((.((((((................)))))).))((((((((.....(((((((.....((((....(((......)))..))))..))))))))))))))). ( -20.19) >DroEre_CAF1 17541 120 - 1 UACAUUCAUAGGCGCUUUUUGUGCAUAUGCAAGACUUAUGUAAUGCACUUGGUUGCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA .............(((((((.(((....))).(((((.((((.((((.((((((((((......((((.......))))...)))))))))).)))))))))))))......))))))). ( -29.80) >DroYak_CAF1 17349 110 - 1 UGCAUU----------UCGUGUGCAUAUGUAACACUUAUGUAAUGCACUUGGUUUCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA ......----------.((.(((((((((((.....))))).)))))).))((((((((.....(((((((.....((((....(((......)))..))))..))))))))))))))). ( -22.70) >consensus UGUAAU__________UCGUGUGCAUAUGCAACACUUAUAUAAUGCACUUGGUUUCUUUAUAUCCAAUUAAAUUCGUUGUCCAAGCAAUUAAAUGCAUGCAAAGUUAAUUGAGAGAAGCA ....................((((((.((((.......))))))))))...((((((((.....(((((((.....((((....(((......)))..))))..))))))))))))))). (-19.00 = -19.44 + 0.44)


| Location | 9,954,983 – 9,955,092 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -19.11 |
| Consensus MFE | -9.59 |
| Energy contribution | -9.27 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9954983 109 + 22224390 ACAACGAAUUUAAUUGGAUAUAAAGAAACCAAGUGCAUUAUAUAAGUGUUACAUAUGCACACGA----------AUUAUAUAUACAUAUAUGCAUAUGCAAUAGGCCAACAAUCUAACA .............((((((((((.(..........).))))))...((((.((((((((.....----------..(((((....))))))))))))).))))..)))).......... ( -13.11) >DroSec_CAF1 17522 100 + 1 ACAACGAAUUUAAUUGGAUAUAAAGAAACCAAGUGCAUUAUAUAAGUGUUGCAUAUGCACACCA----------AUUACAU--------AUGCAUAUGC-AUAGGCCAACAAUCUAACA .............((((((...............(((((.....)))))((((((((((.....----------.......--------.)))))))))-)..........)))))).. ( -17.32) >DroSim_CAF1 4219 100 + 1 ACAACGAAUUUAAUUGGAUAUAAAGAAACCAAGUGCAUUAUAUAAGUGUUGCAUAUGCACACGA----------AUUACAU--------AUGCAUAUGC-AUAGGCCAACAAUCUAACA .............((((((...............(((((.....)))))((((((((((.....----------.......--------.)))))))))-)..........)))))).. ( -17.32) >DroEre_CAF1 17581 118 + 1 ACAACGAAUUUAAUUGGAUAUAAAGCAACCAAGUGCAUUACAUAAGUCUUGCAUAUGCACAAAAAGCGCCUAUGAAUGUAUAUACAUAUAUAUUCAUGC-AUAAGCCAACAAUCCAAUA ............(((((((.....((......((((((..((.......))...)))))).......((..(((((((((((....)))))))))))))-....)).....))))))). ( -27.20) >DroYak_CAF1 17389 102 + 1 ACAACGAAUUUAAUUGGAUAUAAAGAAACCAAGUGCAUUACAUAAGUGUUACAUAUGCACACGA----------AAUGCAUAUACAUAUA------UGC-AUAGGUCAACAAUCUAACA .............((((((........(((..((((((.(((....))).....))))))....----------.((((((((....)))------)))-)).))).....)))))).. ( -20.62) >consensus ACAACGAAUUUAAUUGGAUAUAAAGAAACCAAGUGCAUUAUAUAAGUGUUGCAUAUGCACACGA__________AUUACAUAUACAUAUAUGCAUAUGC_AUAGGCCAACAAUCUAACA .............((((((.............((((((................)))))).....................................((.....)).....)))))).. ( -9.59 = -9.27 + -0.32)


| Location | 9,955,023 – 9,955,114 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.84 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -6.94 |
| Energy contribution | -7.42 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955023 91 + 22224390 UAUAAGUGUUACAUAUGCACACGA----------AUUAUAUAUACAUAUAUGCAUAUGCAAUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGU------------------CAG ......((((.((((((((.....----------..(((((....))))))))))))).))))(((((....((((((......)))).))...))))------------------).. ( -18.51) >DroSec_CAF1 17562 100 + 1 UAUAAGUGUUGCAUAUGCACACCA----------AUUACAU--------AUGCAUAUGC-AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGUCAAGUUUCUGCUGGAAAUCAG .....((.(((((((((((.....----------.......--------.)))))))))-).).))......((((.((((((((((((....)))).)))..))))).))))...... ( -23.32) >DroSim_CAF1 4259 100 + 1 UAUAAGUGUUGCAUAUGCACACGA----------AUUACAU--------AUGCAUAUGC-AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGUUCAAGUUUCUGCUGGAAAUCAG .....((.(((((((((((.....----------.......--------.)))))))))-).).))......((((.((((((((.(((....)))..)))..))))).))))...... ( -22.12) >DroEre_CAF1 17621 118 + 1 CAUAAGUCUUGCAUAUGCACAAAAAGCGCCUAUGAAUGUAUAUACAUAUAUAUUCAUGC-AUAAGCCAACAAUCCAAUAGAAUUGUUAUGAAAAUGGCCAACUUUCUGUUAGAAAUAAG .....((...((.((((((.............((((((((((....)))))))))))))-))).))..))..((.(((((((..(((((....))))).....))))))).))...... ( -23.81) >DroYak_CAF1 17429 102 + 1 CAUAAGUGUUACAUAUGCACACGA----------AAUGCAUAUACAUAUA------UGC-AUAGGUCAACAAUCUAACAGAAUUGUUAUAAAAAUGGCCAACUUUCUGUUCGAAAUCAG .....((((.......)))).(((----------(((((((((....)))------)))-)).(((((......((((......))))......))))).........))))....... ( -22.40) >consensus UAUAAGUGUUGCAUAUGCACACGA__________AUUACAUAUACAUAUAUGCAUAUGC_AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGUCAACUUUCUGCUGGAAAUCAG .....((((.......))))...........................................(((((......((((......))))......))))).................... ( -6.94 = -7.42 + 0.48)



| Location | 9,955,023 – 9,955,114 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.84 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -11.85 |
| Energy contribution | -13.37 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.45 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955023 91 - 22224390 CUG------------------ACCAUUUUCAUAACAAUUCUGUUAGAUUGUUGGCCUAUUGCAUAUGCAUAUAUGUAUAUAUAAU----------UCGUGUGCAUAUGUAACACUUAUA ...------------------.(((...((.(((((....)))))))....)))....((((((((((((((..((.......))----------..))))))))))))))........ ( -21.50) >DroSec_CAF1 17562 100 - 1 CUGAUUUCCAGCAGAAACUUGACCAUUUUCAUAACAAUUCUGUUAGAUUGUUGGCCUAU-GCAUAUGCAU--------AUGUAAU----------UGGUGUGCAUAUGCAACACUUAUA .......(((((((.....(((......)))(((((....)))))..)))))))....(-((((((((((--------((.....----------..)))))))))))))......... ( -26.80) >DroSim_CAF1 4259 100 - 1 CUGAUUUCCAGCAGAAACUUGAACAUUUUCAUAACAAUUCUGUUAGAUUGUUGGCCUAU-GCAUAUGCAU--------AUGUAAU----------UCGUGUGCAUAUGCAACACUUAUA .......(((((((.....((((....))))(((((....)))))..)))))))....(-((((((((((--------(((....----------.))))))))))))))......... ( -30.40) >DroEre_CAF1 17621 118 - 1 CUUAUUUCUAACAGAAAGUUGGCCAUUUUCAUAACAAUUCUAUUGGAUUGUUGGCUUAU-GCAUGAAUAUAUAUGUAUAUACAUUCAUAGGCGCUUUUUGUGCAUAUGCAAGACUUAUG ......(((.(((((((((............(((((((((....)))))))))((((((-(.(((.(((((....))))).))).))))))))))))))))((....)).)))...... ( -31.40) >DroYak_CAF1 17429 102 - 1 CUGAUUUCGAACAGAAAGUUGGCCAUUUUUAUAACAAUUCUGUUAGAUUGUUGACCUAU-GCA------UAUAUGUAUAUGCAUU----------UCGUGUGCAUAUGUAACACUUAUG ......((.((((((..((((.............)))))))))).)).(((((.(.(((-(((------(((((((....)))).----------..))))))))).))))))...... ( -21.72) >consensus CUGAUUUCCAACAGAAACUUGACCAUUUUCAUAACAAUUCUGUUAGAUUGUUGGCCUAU_GCAUAUGCAUAUAUGUAUAUGUAAU__________UCGUGUGCAUAUGCAACACUUAUA ...............................((((((((......)))))))).......(((((((((((...........................))))))))))).......... (-11.85 = -13.37 + 1.52)


| Location | 9,955,053 – 9,955,149 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -4.46 |
| Energy contribution | -4.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9955053 96 + 22224390 UAUACAUAUAUGCAUAUGCAAUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGU------------------CAGAAUUCCAUAAAA-CGAUUCAGUUAAU-UGAUGGCAGC ..........(((....)))....((((.((((.((((.((((((((......((((.------------------.......))))..))-)))))).))))))-)).))))... ( -23.10) >DroSec_CAF1 17592 107 + 1 U--------AUGCAUAUGC-AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGUCAAGUUUCUGCUGGAAAUCAGAAUUCCAUAAAAGCGAUUCAGUUAAUUUGAUGCCAGC (--------((((....))-)))(((...(((..((((.((((((((.(....((((......(((((.(....).)))))..)))).).)))))))).))))..)))..)))... ( -27.00) >DroSim_CAF1 4289 107 + 1 U--------AUGCAUAUGC-AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGUUCAAGUUUCUGCUGGAAAUCAGAAUUCCAUAAAAGCGAUUCAGUUAAUUUGAUGCCAGG (--------((((....))-)))(((...(((..((((.((((((((........((((..(((((.....)))))..))))........)))))))).))))..)))..)))... ( -26.89) >DroEre_CAF1 17661 113 + 1 UAUACAUAUAUAUUCAUGC-AUAAGCCAACAAUCCAAUAGAAUUGUUAUGAAAAUGGCCAACUUUCUGUUAGAAAUAAGAAUUCCGUAAAA-UGUAUCAGUUAAU-UGAAGCAAAC ......(((..((((..((-....))......((.(((((((..(((((....))))).....))))))).)).....))))...)))...-(((.((((....)-))).)))... ( -14.70) >DroYak_CAF1 17459 107 + 1 UAUACAUAUA------UGC-AUAGGUCAACAAUCUAACAGAAUUGUUAUAAAAAUGGCCAACUUUCUGUUCGAAAUCAGAAUUCCAUAAAA-UGUAUCAGUUAGU-UAAGGCAAAG .((((((.((------((.-...(((((......((((......))))......)))))....(((((........)))))...))))..)-)))))........-.......... ( -16.00) >consensus UAUACAUAUAUGCAUAUGC_AUAGGCCAACAAUCUAACAGAAUUGUUAUGAAAAUGGUCAACUUUCUGCUGGAAAUCAGAAUUCCAUAAAA_CGAUUCAGUUAAU_UGAUGCCAGC .......................(((((......((((......))))......)))))......................................................... ( -4.46 = -4.74 + 0.28)

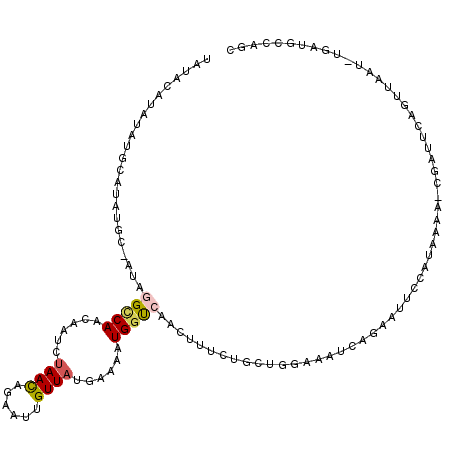
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:16 2006