
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,954,459 – 9,954,688 |
| Length | 229 |
| Max. P | 0.889551 |

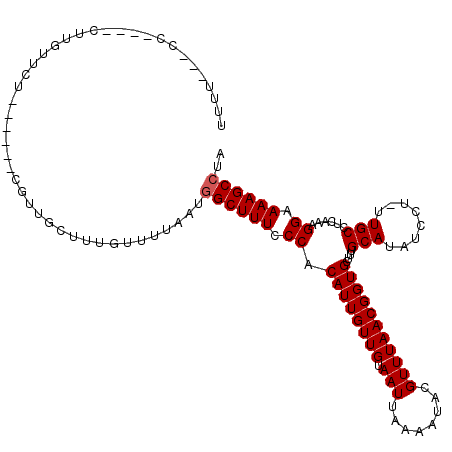
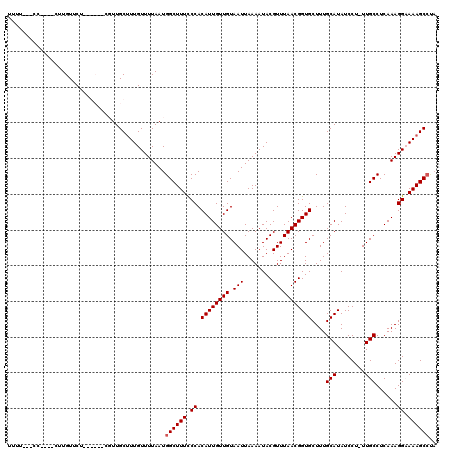
| Location | 9,954,459 – 9,954,579 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
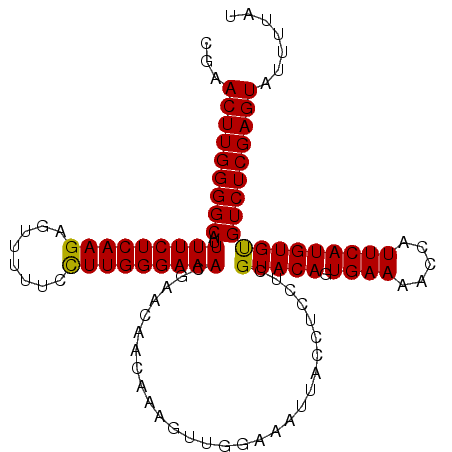
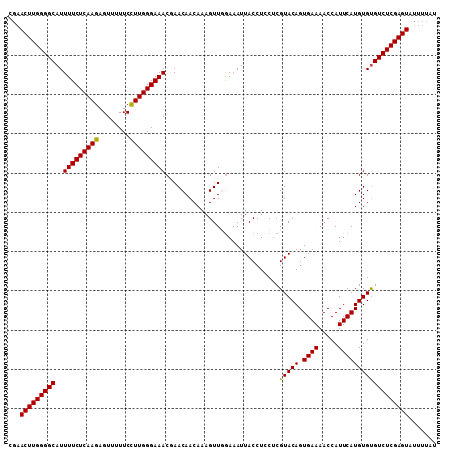
>X_DroMel_CAF1 9954459 120 + 22224390 UUUUUUGCUUUUGCUUGUUCUUUGCUUCGUUGCUUUGUUUUAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCUUUUGCCUCAAAGGCAAAGCCCA ..(((((((((((..........((.((((((...(((((((((((.....))(((....))).)))))))))....)))))).))...(((........)))..))))))))))).... ( -24.50) >DroSec_CAF1 17005 112 + 1 UUUUUUUGC----CUUGUUCUUUGCUUCGUUGCUUUGUUUUAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGCAAAGC--- ...((((((----(((.......((.((((((...(((((((((((.....))(((....))).)))))))))....)))))).))...(((......-.)))....))))))))).--- ( -23.90) >DroEre_CAF1 17091 106 + 1 UUCU---CC----CUUGUUCU------CUUUGCAUUGUUUUAAUGGCUUUUCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGAAAAGCCUA ....---..----........------.................(((((((((.((((((((.(((........)))))))))))....(((......-.)))......))))))))).. ( -21.90) >DroYak_CAF1 16875 104 + 1 UUUU---CC----CUUGUUC--------UUUGCUUUGUUUUAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGAAAAGCCUA ....---..----.......--------................((((((....((((((((.(((........))))))))))).........((((-(((...))))))))))))).. ( -18.40) >consensus UUUU___CC____CUUGUUCU______CGUUGCUUUGUUUUAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU_UUGCCUCAAAGGAAAAGCCUA ............................................((((((.((.((((((((.(((........)))))))))))....(((........)))......)).)))))).. (-16.20 = -16.45 + 0.25)

| Location | 9,954,459 – 9,954,579 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -15.76 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9954459 120 - 22224390 UGGGCUUUGCCUUUGAGGCAAAAGGAUAUGCAAAGCACCGUUAAACGUAUUUUAAUUACAACAAUGUGGGAAAGCCAUUAAAACAAAGCAACGAAGCAAAGAACAAGCAAAAGCAAAAAA ((..((((((.((((..((....((...(((...)))))(((....(((.......)))......((((.....))))...)))...))..))))))))))..)).((....))...... ( -22.90) >DroSec_CAF1 17005 112 - 1 ---GCUUUGCCUUUGAGGCAA-AGGAUAUGCAAAGCACCGUUAAACGUAUUUUAAUUACAACAAUGUGGGAAAGCCAUUAAAACAAAGCAACGAAGCAAAGAACAAG----GCAAAAAAA ---((((((((((((...)))-))).........((..((((....(((.......)))......((((.....))))...........))))..))......))))----))....... ( -20.70) >DroEre_CAF1 17091 106 - 1 UAGGCUUUUCCUUUGAGGCAA-AGGAUAUGCAAAGCACCGUUAAACGUAUUUUAAUUACAACAAUGUGGAAAAGCCAUUAAAACAAUGCAAAG------AGAACAAG----GG---AGAA ......(((((((((..(((.-......)))...(((..(((....(((.......)))......((((.....))))...)))..)))....------....))))----))---))). ( -19.70) >DroYak_CAF1 16875 104 - 1 UAGGCUUUUCCUUUGAGGCAA-AGGAUAUGCAAAGCACCGUUAAACGUAUUUUAAUUACAACAAUGUGGGAAAGCCAUUAAAACAAAGCAAA--------GAACAAG----GG---AAAA ..(((((((((.(((..(((.-......)))...............(((.......)))..)))...)))))))))................--------.......----..---.... ( -18.90) >consensus UAGGCUUUGCCUUUGAGGCAA_AGGAUAUGCAAAGCACCGUUAAACGUAUUUUAAUUACAACAAUGUGGGAAAGCCAUUAAAACAAAGCAAAG______AGAACAAG____GG___AAAA ..(((((((((.(((..(((........)))...............(((.......)))..)))...)))))))))............................................ (-15.76 = -16.45 + 0.69)



| Location | 9,954,499 – 9,954,619 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.05 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9954499 120 + 22224390 UAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCUUUUGCCUCAAAGGCAAAGCCCACGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAA ...(((.....)))((((((((.(((........)))))))))))...(((......(((((((.....)))))))((((......))))))).(((((((((........))))))))) ( -29.30) >DroSec_CAF1 17041 113 + 1 UAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGCAAAGC------ACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAA .....((...((((....((((.(((........)))))))((((((((((.....((-(((...))))))))))))------)))))))))..(((((((((........))))))))) ( -30.20) >DroEre_CAF1 17118 119 + 1 UAAUGGCUUUUCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGAAAAGCCUACGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAA ....(((((((((.((((((((.(((........)))))))))))....(((......-.)))......)))))))))................(((((((((........))))))))) ( -30.40) >DroYak_CAF1 16900 119 + 1 UAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU-UUGCCUCAAAGGAAAAGCCUACGAACUUGGGGCAUUUUCUCAAGAGUUUUUCUUUGGGAAA ........((((((.....(((((.........((.....))..(((((.....((((-(((...)))))))))))).))))).(((((((.....)))))))..........)))))). ( -26.60) >consensus UAAUGGCUUUCCCACAUUGUUGUAAUUAAAAUACGUUUAACGGUGCUUUGCAUAUCCU_UUGCCUCAAAGGAAAAGCCUACGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAA ........(((((.((((((((.(((........)))))))))))....(((........)))....((((((((((.......(((((((.....))))))).))))))))))))))). (-24.92 = -25.05 + 0.13)

| Location | 9,954,579 – 9,954,688 |
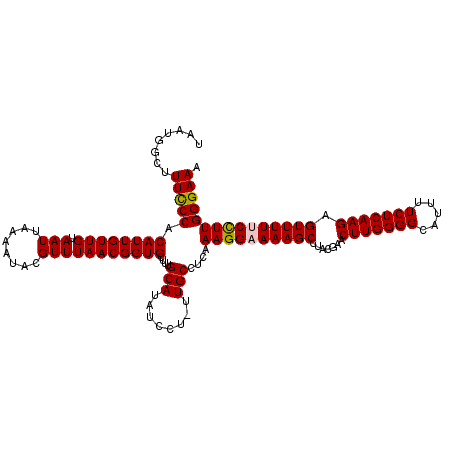
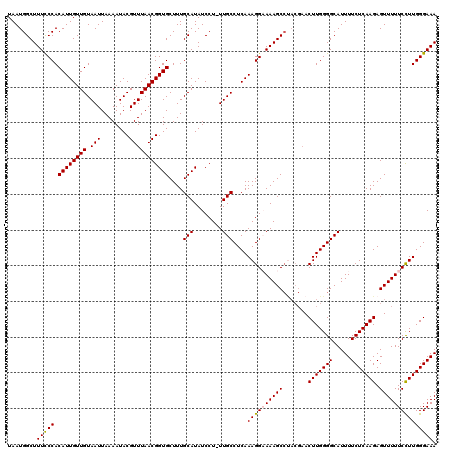
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -24.17 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9954579 109 + 22224390 CGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAACGAACAACAAAGUUGAAAAUUACCUUCUCGUACAGUGAAAACCAUUCAUGUGUGUCUCGAGUAUUUUAU ...((((((((((((((((((((........)))))))))....((((...))))................(((.((((.....))))))))))))))))))....... ( -25.60) >DroSec_CAF1 17117 106 + 1 ---ACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAACAAACAACAAAGUUGGAAAUUACCUACACGUACAGUGAAAACCAUUCAUGUGUGUCUCGAGUAUUUUAU ---(((((((((..(((((((((........)))))))))....((((...))))..........((((((..((((.....)))).)))))))))))))))....... ( -27.80) >DroEre_CAF1 17197 109 + 1 CGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAACUAACAACAAAGUUUGAAAUUACCUCCUCGUACAGUGAAAACCAUUCAUGUGCGUCUCGAGUAUUUUAU ...(((((((((..(((((((((........))))))))).............................(((((.((((.....))))))))))))))))))....... ( -28.70) >DroYak_CAF1 16979 109 + 1 CGAACUUGGGGCAUUUUCUCAAGAGUUUUUCUUUGGGAAACGAACAACAAAGUUUGAAAUUACCUCCUCGUACAGUGAAAACCAUUCAUGUGUGUCUCGAGUAUUUUAU ...((((((((((((((((((((........)))))))))(((((......)))))...............(((.((((.....))))))))))))))))))....... ( -25.50) >consensus CGAACUUGGGGCAUUUUCUCAAGAGUUUUUCCUUGGGAAACGAACAACAAAGUUGGAAAUUACCUCCUCGUACAGUGAAAACCAUUCAUGUGUGUCUCGAGUAUUUUAU ...(((((((((..(((((((((........))))))))).............................(((((.((((.....))))))))))))))))))....... (-24.17 = -23.80 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:11 2006