
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,953,100 – 9,953,245 |
| Length | 145 |
| Max. P | 0.999171 |

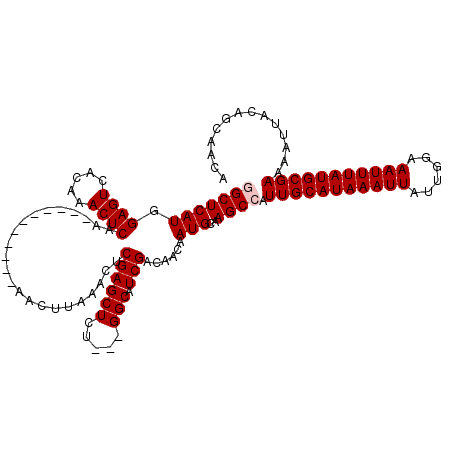
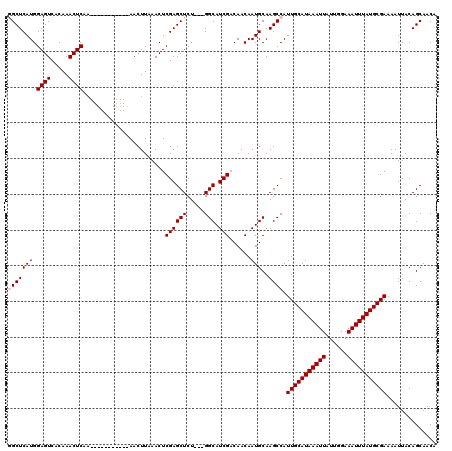
| Location | 9,953,100 – 9,953,206 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9953100 106 + 22224390 GGCUCAUGGAGUCACAAACUCAA-----------AACUUAAACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA (((((((.((((.....))))..-----------.........((((((...---.)).)))).....)))..)))).(((((((((((......))))))))))).............. ( -24.60) >DroSec_CAF1 15717 106 + 1 GGCUCAUGGAGUCACAAACUCAA-----------AACUUAAACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA (((((((.((((.....))))..-----------.........((((((...---.)).)))).....)))..)))).(((((((((((......))))))))))).............. ( -24.60) >DroSim_CAF1 1868 106 + 1 GGCUCAUGGAGUCACAAACUCAA-----------AACUUAAACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA (((((((.((((.....))))..-----------.........((((((...---.)).)))).....)))..)))).(((((((((((......))))))))))).............. ( -24.60) >DroEre_CAF1 15694 120 + 1 AGCUCAUGGAGUCACAAACUCAAACUCAAACUCAAACUUAAACUCGAGCUCUCGUGGCAUCGUCAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA .(((((((((((.....)))).........(((............)))....))))((((.(....).))))......(((((((((((......))))))))))).......))).... ( -22.20) >DroYak_CAF1 15476 105 + 1 AGCUCAUGGAGUCACAAACUC------------CAACUUAAACUCGAGCUCU---GGCAUCGUCAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA .(((..((((((.....))))------------))..........(.(((..---.((((.(....).)))).)))).(((((((((((......))))))))))).......))).... ( -26.20) >consensus GGCUCAUGGAGUCACAAACUCAA___________AACUUAAACUCGAGCUCU___GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACA (((((((.((((.....)))).......................((((((.....))).)))......)))..)))).(((((((((((......))))))))))).............. (-20.46 = -20.86 + 0.40)

| Location | 9,953,100 – 9,953,206 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9953100 106 - 22224390 UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUUUAAGUU-----------UUGAGUUUGUGACUCCAUGAGCC ................(((((((((......)))))))))...(((((((((((....)))))).---.(((((((((.......)-----------))))))))..........))))) ( -28.90) >DroSec_CAF1 15717 106 - 1 UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUUUAAGUU-----------UUGAGUUUGUGACUCCAUGAGCC ................(((((((((......)))))))))...(((((((((((....)))))).---.(((((((((.......)-----------))))))))..........))))) ( -28.90) >DroSim_CAF1 1868 106 - 1 UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUUUAAGUU-----------UUGAGUUUGUGACUCCAUGAGCC ................(((((((((......)))))))))...(((((((((((....)))))).---.(((((((((.......)-----------))))))))..........))))) ( -28.90) >DroEre_CAF1 15694 120 - 1 UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGACGAUGCCACGAGAGCUCGAGUUUAAGUUUGAGUUUGAGUUUGAGUUUGUGACUCCAUGAGCU ....(((.........(((((((((......))))))))).((((...((((((....))))))(((((..((((((..(((((......)))))..)))))))))))...)))).))). ( -34.30) >DroYak_CAF1 15476 105 - 1 UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGACGAUGCC---AGAGCUCGAGUUUAAGUUG------------GAGUUUGUGACUCCAUGAGCU ....(((......((((((((((((......)))))))))...(((((((((((....)))))).---.))))).))).......((------------(((((...)))))))..))). ( -31.00) >consensus UGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC___AGAGCUCGAGUUUAAGUU___________UUGAGUUUGUGACUCCAUGAGCC ....(((......((((((((((((......)))))))))...(((((((((((....)))))).....))))).)))....)))................(((..((....))..))). (-25.12 = -25.12 + -0.00)

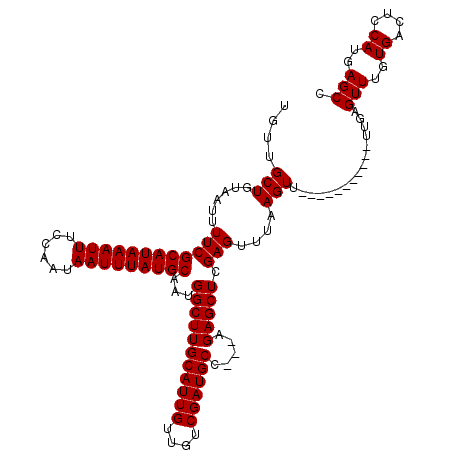
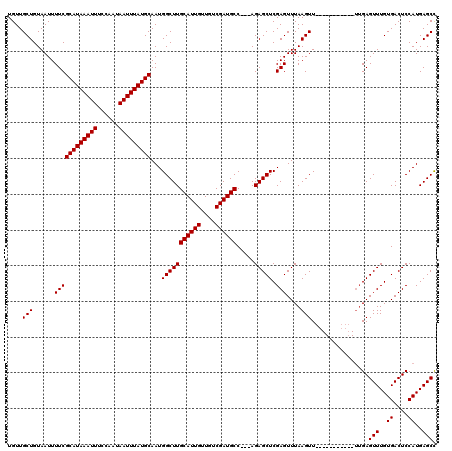
| Location | 9,953,129 – 9,953,245 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.52 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9953129 116 + 22224390 AACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACUGCGACGGCAAAACGACC-ACAAUGUCAGCAACGGGAA ..((((.((...---.))...........(((..(((.(((((((((((......)))))))))))........(((.....)))...))).....(((.-.....))).))).)))).. ( -29.90) >DroSec_CAF1 15746 117 + 1 AACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACUGCGACGGCAAAACGACCAACAAUGUCAGCAACGGGAA ..((((.((...---.))...........(((..(((.(((((((((((......)))))))))))........(((.....)))...))).....(((.......))).))).)))).. ( -29.40) >DroSim_CAF1 1897 117 + 1 AACUCGAGCUCU---GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACUGCGACGGCAAAACGACCAACAAUGUCAGCAACGGGAA ..((((.((...---.))...........(((..(((.(((((((((((......)))))))))))........(((.....)))...))).....(((.......))).))).)))).. ( -29.40) >DroEre_CAF1 15734 119 + 1 AACUCGAGCUCUCGUGGCAUCGUCAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACAGCGACAGCAAAACGACC-ACAAUGUCGGCAACGGGAA ..((((.(((.((((.((((.(....).))))......(((((((((((......))))))))))).................)))).)))....((((.-.....))))....)))).. ( -31.30) >DroYak_CAF1 15504 116 + 1 AACUCGAGCUCU---GGCAUCGUCAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACUGCGACAGCAAAACGACC-ACAAUGUCUGCAACGGGAA ..((((.(((..---.((((.(....).)))).)))..(((((((((((......)))))))))))........(((.....(((....)))....(((.-.....))))))..)))).. ( -27.50) >consensus AACUCGAGCUCU___GGCAUCGACAACAAUGCAAGCCAUUGCAUAAAUUAUUGGAAAUUUAUGCGAAAAUUACAGCAACAACUGCGACGGCAAAACGACC_ACAAUGUCAGCAACGGGAA ..((((.(((.....)))...........(((..(((.(((((((((((......)))))))))))........(((.....)))...))).....(((.......))).))).)))).. (-26.08 = -26.68 + 0.60)



| Location | 9,953,129 – 9,953,245 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.52 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9953129 116 - 22224390 UUCCCGUUGCUGACAUUGU-GGUCGUUUUGCCGUCGCAGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUU ....((..((((((.....-.))).....(((((.(((((....))))).......(((((((((......))))))))).)))))..((((((....)))))).---..))).)).... ( -32.00) >DroSec_CAF1 15746 117 - 1 UUCCCGUUGCUGACAUUGUUGGUCGUUUUGCCGUCGCAGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUU ...((((((((((.((((((((..(((((((.((..((((....))))..))....))).))))..))))))))))).)))))))((.((((((....)))))).---)).......... ( -34.60) >DroSim_CAF1 1897 117 - 1 UUCCCGUUGCUGACAUUGUUGGUCGUUUUGCCGUCGCAGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC---AGAGCUCGAGUU ...((((((((((.((((((((..(((((((.((..((((....))))..))....))).))))..))))))))))).)))))))((.((((((....)))))).---)).......... ( -34.60) >DroEre_CAF1 15734 119 - 1 UUCCCGUUGCCGACAUUGU-GGUCGUUUUGCUGUCGCUGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGACGAUGCCACGAGAGCUCGAGUU ....(((((.(((((.(((-(((((((((((.((.((....)).)).)))).....(((((((((......))))))))))))))).)))).))))).)))))...(((....))).... ( -32.30) >DroYak_CAF1 15504 116 - 1 UUCCCGUUGCAGACAUUGU-GGUCGUUUUGCUGUCGCAGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGACGAUGCC---AGAGCUCGAGUU .....((((((((((((((-((..(.....)..)))))).)))..))))))).((((((((((((......)))))))))...(((((((((((....)))))).---.))))).))).. ( -33.30) >consensus UUCCCGUUGCUGACAUUGU_GGUCGUUUUGCCGUCGCAGUUGUUGCUGUAAUUUUCGCAUAAAUUUCCAAUAAUUUAUGCAAUGGCUUGCAUUGUUGUCGAUGCC___AGAGCUCGAGUU ....(((((.(((((.(((.(((((((........(((((....))))).......(((((((((......)))))))))))))))).))).))))).)))))................. (-30.62 = -30.90 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:07 2006