
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,952,527 – 9,952,684 |
| Length | 157 |
| Max. P | 0.925769 |

| Location | 9,952,527 – 9,952,644 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9952527 117 + 22224390 AAUGAUCAUUGGAAAUUCUUAAAAAUUUUCUUGGGAAAAUCGCGUGCCUCACGCGUGAAUAUCGAAUGAGUACACAACUUGCCAUCGAUUCAUUACGAUACCAAGUUGAAGUCGAGA ..((((((((.((.((((......((((((....))))))((((((...)))))).)))).)).))))).....(((((((..((((........))))..)))))))...)))... ( -31.10) >DroSec_CAF1 15139 117 + 1 AAUGAUCAUUGGAAAUUCUUUAAAAUUUCCUUGCGAAAAUCGCGUGCCUCACGCGUGAGUAUCGAAUGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUGAAGUCGGGG (((((((...(((((((......)))))))..((((...(((((((.....)))))))...))....((((.....))))))....)).))))).(((((.(.....).)))))... ( -28.10) >DroEre_CAF1 15133 117 + 1 AAUUAUCAUUGAAAAUUCUUAAACAUUUUCUUGGGAAAAUCGCGUGCCCCACGCGUGAAUAUCGAAUGAGUACACAACUUGCCAUCGAUUCAUUACGACUGAGAGUUAAAGUCAAGA ........((((.((((((((...((((((....)))))).(((((...)))))((((..(((((.((.(((.......))))))))))...))))...))))))))....)))).. ( -24.70) >DroYak_CAF1 14912 117 + 1 GAUUAUCAUUGAAAAUUCUUUAAAAUUUCCUUGGGAAAAUCGCGUGCCUCACGCGUGAAUAUCGAACGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUAAUGCCAUGA ........................(((..(((((((.....(((((...)))))((((..(((((.(((((.....)))))...)))))...))))..)))))))..)))....... ( -28.50) >consensus AAUGAUCAUUGAAAAUUCUUAAAAAUUUCCUUGGGAAAAUCGCGUGCCUCACGCGUGAAUAUCGAAUGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUAAAGUCAAGA .........................((..(((((...(((((((((...)))))((((..(((((.(((((.....)))))...)))))...)))))))))))))..))........ (-20.98 = -21.35 + 0.37)


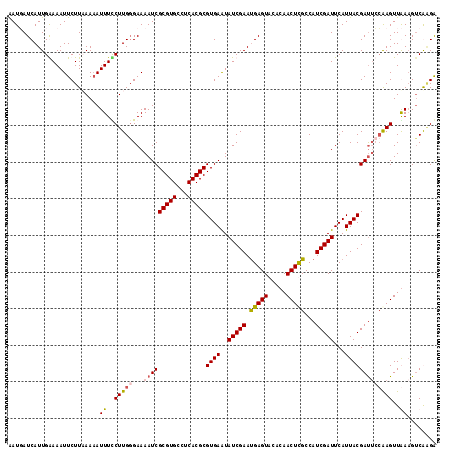
| Location | 9,952,527 – 9,952,644 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -28.19 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
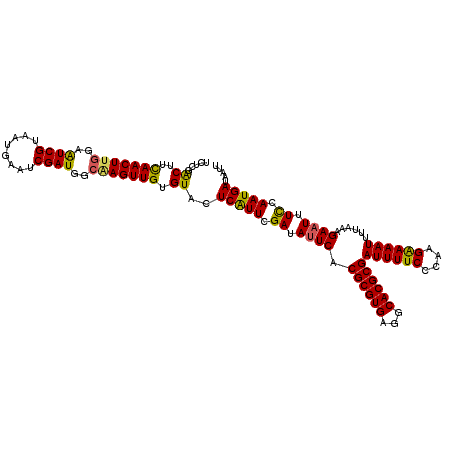
>X_DroMel_CAF1 9952527 117 - 22224390 UCUCGACUUCAACUUGGUAUCGUAAUGAAUCGAUGGCAAGUUGUGUACUCAUUCGAUAUUCACGCGUGAGGCACGCGAUUUUCCCAAGAAAAUUUUUAAGAAUUUCCAAUGAUCAUU .....((..(((((((.(((((........))))).))))))).))..(((((.((.((((.((((((...))))))((((((....))))))......)))).)).)))))..... ( -33.90) >DroSec_CAF1 15139 117 - 1 CCCCGACUUCAACUUGGAAUCGUAAUGAAUCGAUGGCGAGUUGUGUACUCAUUCGAUACUCACGCGUGAGGCACGCGAUUUUCGCAAGGAAAUUUUAAAGAAUUUCCAAUGAUCAUU ...(((.(((.....))).))).(((((.(((...(((((((((((.(((((.((.......)).)))))..)))))))..))))..((((((((....))))))))..)))))))) ( -33.40) >DroEre_CAF1 15133 117 - 1 UCUUGACUUUAACUCUCAGUCGUAAUGAAUCGAUGGCAAGUUGUGUACUCAUUCGAUAUUCACGCGUGGGGCACGCGAUUUUCCCAAGAAAAUGUUUAAGAAUUUUCAAUGAUAAUU ....((((.........)))).......((((.(((.(((((((((.(((((.((.......)).)))))..)))))))))..))).((((((........))))))..)))).... ( -24.60) >DroYak_CAF1 14912 117 - 1 UCAUGGCAUUAACUUGGAAUCGUAAUGAAUCGAUGGCGAGUUGUGUACUCGUUCGAUAUUCACGCGUGAGGCACGCGAUUUUCCCAAGGAAAUUUUAAAGAAUUUUCAAUGAUAAUC ((((........(((((........((((((((..((((((.....)))))))))))..)))((((((...))))))......)))))(((((((....)))))))..))))..... ( -30.70) >consensus UCUCGACUUCAACUUGGAAUCGUAAUGAAUCGAUGGCAAGUUGUGUACUCAUUCGAUAUUCACGCGUGAGGCACGCGAUUUUCCCAAGAAAAUUUUAAAGAAUUUCCAAUGAUAAUU .....((..(((((((..((((........))))..))))))).))..(((((.((.((((.((((((...))))))((((((....))))))......)))).)).)))))..... (-28.19 = -27.50 + -0.69)

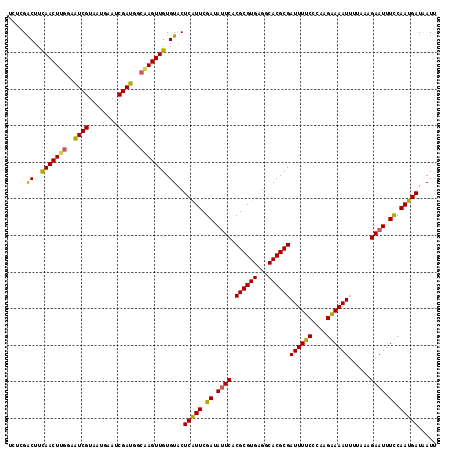
| Location | 9,952,567 – 9,952,684 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
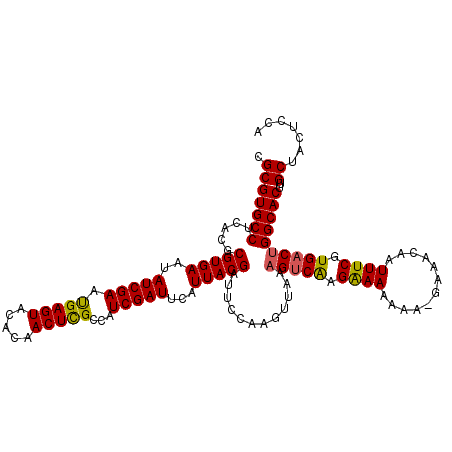
>X_DroMel_CAF1 9952567 117 + 22224390 CGCGUGCCUCACGCGUGAAUAUCGAAUGAGUACACAACUUGCCAUCGAUUCAUUACGAUACCAAGUUGAAGUCGAGAAAAAAAGAAAACAAUUUCGUGACUGGCACUUGCUACUCCA ((((((...))))))............((((...(((((((..((((........))))..))))))).(((((.((((............)))).)))))(((....))))))).. ( -29.10) >DroSec_CAF1 15179 115 + 1 CGCGUGCCUCACGCGUGAGUAUCGAAUGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUGAAGUCGGGGAAAAA--GAAACAAUUUCGUGACUGGCACUUGCUACUCCA .(((((((((((((((((..(((((.(((((.....)))))...)))))...))))).((((..(.......)..))))...--..........)))))..)))))..))....... ( -31.80) >DroEre_CAF1 15173 117 + 1 CGCGUGCCCCACGCGUGAAUAUCGAAUGAGUACACAACUUGCCAUCGAUUCAUUACGACUGAGAGUUAAAGUCAAGAAAAAAAUGAAACAAUUUCGUGACUGGCACUUGCUACUCCA .(((((((.((..(((((..(((((.((.(((.......))))))))))...)))))..))........(((((.((((............)))).))))))))))..))....... ( -26.30) >DroYak_CAF1 14952 116 + 1 CGCGUGCCUCACGCGUGAAUAUCGAACGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUAAUGCCAUGAAAAAAA-GAAACAAUUUCGUGACUGGCACUUGCUACUCCA .(((((((((((((((((..(((((.(((((.....)))))...)))))...))))).(((...((....))...))).....-..........)))))..)))))..))....... ( -28.50) >consensus CGCGUGCCUCACGCGUGAAUAUCGAAUGAGUACACAACUCGCCAUCGAUUCAUUACGAUUCCAAGUUAAAGUCAAGAAAAAAA_GAAACAAUUUCGUGACUGGCACUUGCUACUCCA .(((((((.....(((((..(((((.(((((.....)))))...)))))...)))))............(((((.((((............)))).))))))))))..))....... (-25.80 = -25.43 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:04 2006