
| Sequence ID | X_DroMel_CAF1 |
|---|---|
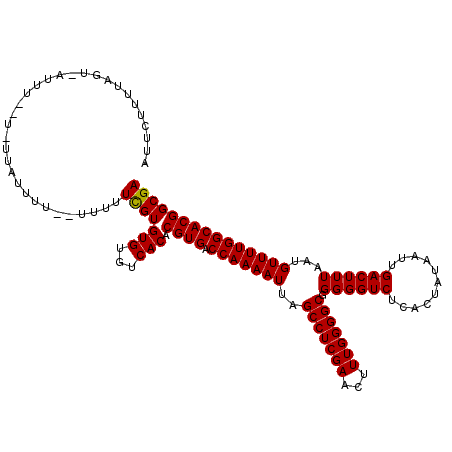
| Location | 9,952,274 – 9,952,390 |
| Length | 116 |
| Max. P | 0.972935 |

| Location | 9,952,274 – 9,952,390 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -28.02 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9952274 116 + 22224390 AUUCUUUUAGUGAUUUAUU----UUUUAUUUUUUCGUGUGUGUCACACGUGACCAAAAUUAGCCUCGAACUUUGGGGCGGGGGUCUCACUUUAAUUGACUUUAAUGUUUUGGCACGGCGA .........(((((.(((.----..............))).))))).((((.(((((((..(((((((...))))))).((((((...........))))))...))))))))))).... ( -29.76) >DroEre_CAF1 14896 103 + 1 AUUCUUUU------------UUAUU-----UUUUCGUGUGUGUCACACGUGACCAAAAUUAGCCUCGAACUUUGGGGCGGGGGUCUCAUUAUAAUUGACUUUAAUGUUUUGGCACGGCGA ........------------.....-----...(((((((...))).((((.(((((((..(((((((...))))))).((((((...........))))))...))))))))))))))) ( -28.50) >DroYak_CAF1 14651 120 + 1 AUUCUUUUAGUAAUUUUCUUUUAUUUUUAUUUUUUGUGUGUGUCACACGUGACCAAAAUUAGCCUCGAACUUUGGGGCGGGGGUCUCACUAUAAUUGACUUUAAUGUUUUGGCACGGCGA .........((.......................((((.....))))((((.(((((((..(((((((...))))))).((((((...........))))))...))))))))))))).. ( -28.10) >consensus AUUCUUUUAGU_AUUU__U_UUAUUUU__UUUUUCGUGUGUGUCACACGUGACCAAAAUUAGCCUCGAACUUUGGGGCGGGGGUCUCACUAUAAUUGACUUUAAUGUUUUGGCACGGCGA .................................(((((((...))).((((.(((((((..(((((((...))))))).((((((...........))))))...))))))))))))))) (-28.02 = -27.80 + -0.22)

| Location | 9,952,274 – 9,952,390 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9952274 116 - 22224390 UCGCCGUGCCAAAACAUUAAAGUCAAUUAAAGUGAGACCCCCGCCCCAAAGUUCGAGGCUAAUUUUGGUCACGUGUGACACACACGAAAAAAUAAAA----AAUAAAUCACUAAAAGAAU ((((((((((((((.......(((...........)))....(((.(.......).)))...)))))).)))).))))...................----................... ( -19.90) >DroEre_CAF1 14896 103 - 1 UCGCCGUGCCAAAACAUUAAAGUCAAUUAUAAUGAGACCCCCGCCCCAAAGUUCGAGGCUAAUUUUGGUCACGUGUGACACACACGAAAA-----AAUAA------------AAAAGAAU (((((((((((((((((((..........)))))........(((.(.......).)))...)))))).)))).))))............-----.....------------........ ( -20.20) >DroYak_CAF1 14651 120 - 1 UCGCCGUGCCAAAACAUUAAAGUCAAUUAUAGUGAGACCCCCGCCCCAAAGUUCGAGGCUAAUUUUGGUCACGUGUGACACACACAAAAAAUAAAAAUAAAAGAAAAUUACUAAAAGAAU (((((((((((((((((((..........)))))........(((.(.......).)))...)))))).)))).)))).......................................... ( -20.20) >consensus UCGCCGUGCCAAAACAUUAAAGUCAAUUAUAGUGAGACCCCCGCCCCAAAGUUCGAGGCUAAUUUUGGUCACGUGUGACACACACGAAAAA__AAAAUAA_A__AAAU_ACUAAAAGAAU ((((((((((((((.......(((...........)))....(((.(.......).)))...)))))).)))).)))).......................................... (-19.90 = -19.90 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:01 2006