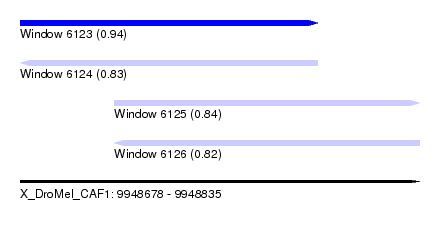
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,948,678 – 9,948,835 |
| Length | 157 |
| Max. P | 0.938508 |

| Location | 9,948,678 – 9,948,795 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.90 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9948678 117 + 22224390 CAAGGCACUCGGCUAUGCCAUU---UCUUUUGUGUUUUUUAAUAAACAAUUAAAAAACCAUUUGGCAGCGCGACAUGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGA ...(((.....))).((((...---........((((((((((.....))))))))))((((((..((((((....(((.((((....)))))))..)))).))..))))))..)))).. ( -38.40) >DroSec_CAF1 11563 117 + 1 CAAGGCGUUCGGCUAUGCCAAU---UCUUUUGUGUUUUUUAAUAAACAAUUAAAAAACCAUUUGGCACCGUGACAUGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGA ....((.(((((...((((((.---.....((.((((((((((.....)))))))))))).))))))))((((((((((.((((....)))))))...)))..))))....))).))... ( -38.00) >DroEre_CAF1 11412 117 + 1 CAAGGCACUCUGCAAAUCUGUU---UUUGUUUUGUUUUUUAAUAAACAAUUAAAAACCCAUUUGGCUGCGGGACAAGAGAGGGGAAUCCCCCCGCCACGUGACUCACAAAUGAAGGCAGA ........(((((......(((---((((..((((((......)))))).))))))).(((((((..(((((......(.((((....))))).)).)))..)...))))))...))))) ( -29.50) >DroYak_CAF1 11094 118 + 1 CAUGGCACUCGGCAAAGCUGUUGCCUUUGGUU--UUUUUUAACAAACAAUUAAAAAACCAUUUGGCUGCGCGAACAGCGAGGGGAUUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGA .........((((...))))((((((((((((--((((.............))))))))(((((...(((((...((((.((((....)))))))).)))).)...))))))))))))). ( -38.62) >consensus CAAGGCACUCGGCAAAGCCAUU___UCUGUUGUGUUUUUUAAUAAACAAUUAAAAAACCAUUUGGCAGCGCGACAAGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGA ....((.....))...(((..............((((((((((.....))))))))))((((((....((((....(((.((((....)))))))..)))).....))))))..)))... (-24.77 = -25.90 + 1.13)


| Location | 9,948,678 – 9,948,795 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9948678 117 - 22224390 UCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCGCGCUGCCAAAUGGUUUUUUAAUUGUUUAUUAAAAAACACAAAAGA---AAUGGCAUAGCCGAGUGCCUUG (((.....((((((..((((((((.((((((((...)))))))))))).).)))..))))))((((((((((.....)))))))))).....)))---...(((((......)))))... ( -39.30) >DroSec_CAF1 11563 117 - 1 UCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCACGGUGCCAAAUGGUUUUUUAAUUGUUUAUUAAAAAACACAAAAGA---AUUGGCAUAGCCGAACGCCUUG ...((.(((........((.((((.((((((((...)))))))))))).)).(((((((.((((((((((((.....)))))))))).)).....---.)))))))....))).)).... ( -37.60) >DroEre_CAF1 11412 117 - 1 UCUGCCUUCAUUUGUGAGUCACGUGGCGGGGGGAUUCCCCUCUCUUGUCCCGCAGCCAAAUGGGUUUUUAAUUGUUUAUUAAAAAACAAAACAAA---AACAGAUUUGCAGAGUGCCUUG ...((.....((((..((((...((((.(.(((((...........))))).).)))).....((((((..((((((......))))))...)))---))).))))..))))..)).... ( -31.60) >DroYak_CAF1 11094 118 - 1 UCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAAUCCCCUCGCUGUUCGCGCAGCCAAAUGGUUUUUUAAUUGUUUGUUAAAAAA--AACCAAAGGCAACAGCUUUGCCGAGUGCCAUG .............(((.(.(((.((((((((((...)))))))))).(((.((.......((((((((((((.....))))..)))--)))))(((((....))))))))))))))))). ( -37.10) >consensus UCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCGCGCAGCCAAAUGGUUUUUUAAUUGUUUAUUAAAAAACAAAAAAAA___AACAGCAUAGCCGAGUGCCUUG ((.((...((((((...((..((((((((((((...)))))))))....)))..))))))))((((((((((.....))))))))))....................)).))........ (-25.27 = -26.40 + 1.13)

| Location | 9,948,715 – 9,948,835 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9948715 120 + 22224390 AAUAAACAAUUAAAAAACCAUUUGGCAGCGCGACAUGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGAAAACUGAAGCUAAAAAUAAUUAUAAAAUGUUCAGCAAAAA ....((((..........((((((..((((((....(((.((((....)))))))..)))).))..))))))....(((....))).....................))))......... ( -26.30) >DroSec_CAF1 11600 120 + 1 AAUAAACAAUUAAAAAACCAUUUGGCACCGUGACAUGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGAAAACUGAAGCUAAAAAUAAUUAUAAGAUGUUCAGCAAAAA ....................((((((.((((.((..(((.((((....)))))))...)).))(((....))).))(((....)))..)))))).......................... ( -23.90) >DroEre_CAF1 11449 120 + 1 AAUAAACAAUUAAAAACCCAUUUGGCUGCGGGACAAGAGAGGGGAAUCCCCCCGCCACGUGACUCACAAAUGAAGGCAGAAAACUGAAGCUAAAAAUAAUUAUAAGAUAUUCAGCAAAAA ....................(((((((.(((.........((((.....))))(((.(((.........)))..)))......))).))))))).......................... ( -22.50) >DroYak_CAF1 11132 120 + 1 AACAAACAAUUAAAAAACCAUUUGGCUGCGCGAACAGCGAGGGGAUUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGAAAACCGAAGCUAAAAAUAAUUAUAAGAUAUUCAGCAAAAA .......(((((......((((((...(((((...((((.((((....)))))))).)))).)...))))))..(((...........))).....)))))................... ( -25.60) >consensus AAUAAACAAUUAAAAAACCAUUUGGCAGCGCGACAAGCGAGGGGAAUCCCCCCGCUACGUGACUCACAAAUGAAGGCAGAAAACUGAAGCUAAAAAUAAUUAUAAGAUAUUCAGCAAAAA .......(((((......((((((....((((....(((.((((....)))))))..)))).....))))))..(((...........))).....)))))................... (-19.62 = -20.00 + 0.38)

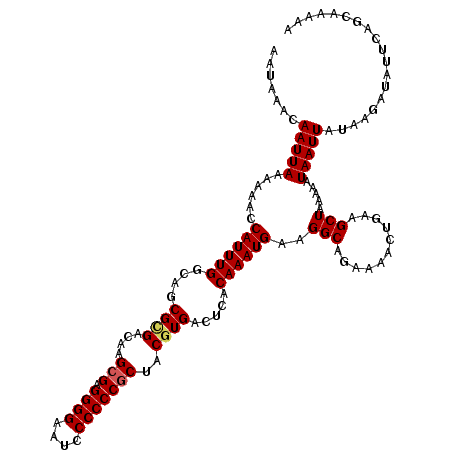
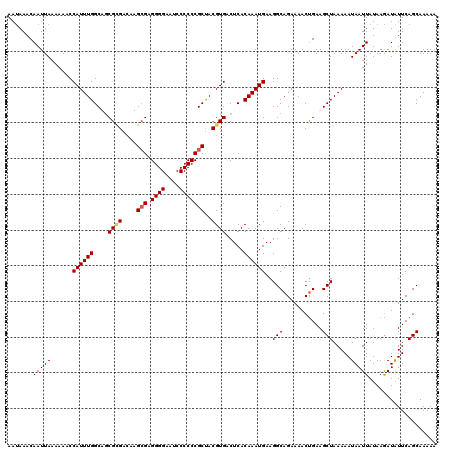
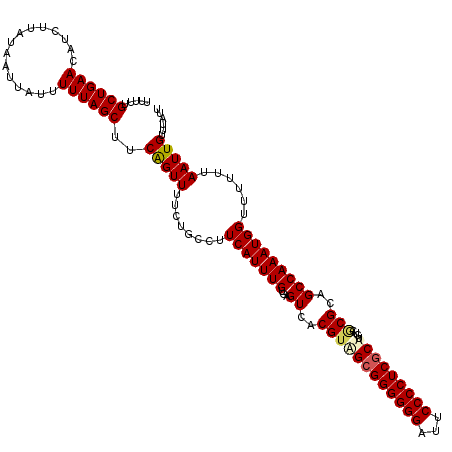
| Location | 9,948,715 – 9,948,835 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -23.85 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9948715 120 - 22224390 UUUUUGCUGAACAUUUUAUAAUUAUUUUUAGCUUCAGUUUUCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCGCGCUGCCAAAUGGUUUUUUAAUUGUUUAUU .......((((((.((...((((((((.((((..(((....))).............((.((((.((((((((...)))))))))))).))))))..))))))))....)).)))))).. ( -31.30) >DroSec_CAF1 11600 120 - 1 UUUUUGCUGAACAUCUUAUAAUUAUUUUUAGCUUCAGUUUUCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCACGGUGCCAAAUGGUUUUUUAAUUGUUUAUU .....((((((...............))))))..(((((........(((((((.(.((.((((.((((((((...)))))))))))).))....))))))))......)))))...... ( -29.40) >DroEre_CAF1 11449 120 - 1 UUUUUGCUGAAUAUCUUAUAAUUAUUUUUAGCUUCAGUUUUCUGCCUUCAUUUGUGAGUCACGUGGCGGGGGGAUUCCCCUCUCUUGUCCCGCAGCCAAAUGGGUUUUUAAUUGUUUAUU .....((((((...............))))))..(((((....(((((((....)))).((..((((.(.(((((...........))))).).))))..)))))....)))))...... ( -27.06) >DroYak_CAF1 11132 120 - 1 UUUUUGCUGAAUAUCUUAUAAUUAUUUUUAGCUUCGGUUUUCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAAUCCCCUCGCUGUUCGCGCAGCCAAAUGGUUUUUUAAUUGUUUGUU ...(((((((((..(((((((........(((....)))............)))))))......(((((((((...)))))))))))))).)))).((((..(((....)))..)))).. ( -32.85) >consensus UUUUUGCUGAACAUCUUAUAAUUAUUUUUAGCUUCAGUUUUCUGCCUUCAUUUGUGAGUCACGUAGCGGGGGGAUUCCCCUCGCAUGUCGCGCAGCCAAAUGGUUUUUUAAUUGUUUAUU .....((((((...............))))))..(((((........(((((((...((..((((((((((((...)))))))))....)))..)))))))))......)))))...... (-23.85 = -24.47 + 0.62)

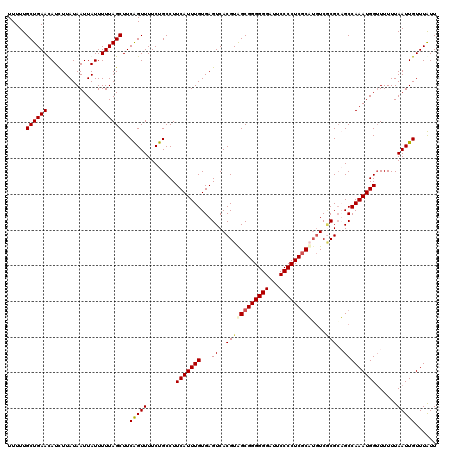
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:58 2006