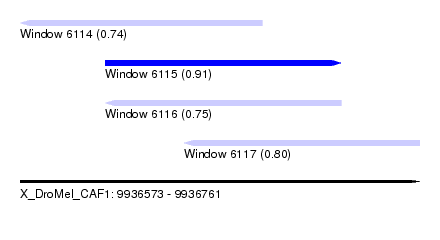
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,936,573 – 9,936,761 |
| Length | 188 |
| Max. P | 0.908035 |

| Location | 9,936,573 – 9,936,687 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -13.20 |
| Consensus MFE | -12.98 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9936573 114 - 22224390 ---AAUAAAAAUACAAAAAAAAAAACCAUCCACAACCACAACAA---CAAGCGGGAGCGGCGAUAAAAUUGUUUUUCAUCUCAUUAUUGCACAAGCGGUCAAAGUAAUUCAUAACACACA ---.....................(((........((.(.....---...).))..((.(((((((...((.....)).....)))))))....)))))..................... ( -11.80) >DroSec_CAF1 6349 114 - 1 AAAAAUAAAAAUACAAAA---ACAACCAUCCACAACCACAACAA---CAAGCGGGAGCGGCGAUAAAAUUGUUUUUCAUCUCAUUAUUGCACAAGUGGUCAAAGUAAUUCAUAACACACA ..................---.............(((((.....---...((....)).(((((((...((.....)).....)))))))....)))))..................... ( -15.50) >DroEre_CAF1 6146 109 - 1 AAA---GAAAAUA--------AAAACAAUCCCCAACCACAACAACACCAAGCGGGAGCAGCGAUAAAAUUGUUUUUCAUCUCAUUAUUGCACAAGUGGUCAAAGUAAUUCAUAACACACA ...---.......--------.............(((((...........((....)).(((((((...((.....)).....)))))))....)))))..................... ( -15.70) >DroYak_CAF1 6540 97 - 1 AAA------------------ACAACCAUACACAAUCACAACAA---CAAGUAGGAGCAGCGAUAAAAUUGUUUUUCAUCUCAUUAUUGCACAAGUGGUCAAAGUAAUUCAUAACA--CA ...------------------.......(((...(((((.....---...((....)).(((((((...((.....)).....)))))))....)))))....)))..........--.. ( -9.80) >consensus AAA___AAAAAUA________AAAACCAUCCACAACCACAACAA___CAAGCGGGAGCAGCGAUAAAAUUGUUUUUCAUCUCAUUAUUGCACAAGUGGUCAAAGUAAUUCAUAACACACA ..................................(((((...........((....)).(((((((...((.....)).....)))))))....)))))..................... (-12.98 = -12.85 + -0.13)


| Location | 9,936,613 – 9,936,724 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.31 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
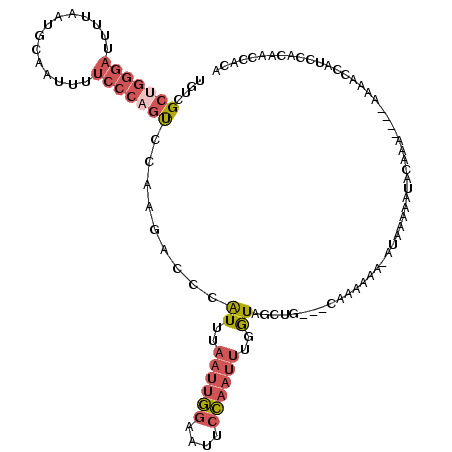
>X_DroMel_CAF1 9936613 111 + 22224390 GAUGAAAAACAAUUUUAUCGCCGCUCCCGCUUG---UUGUUGUGGUUGUGGAUGGUUUUUUUUUUUGUAUUUUUAUU---UUUGCAGCUACCAAAUUGGAAUUCCAAUUAAAUGGGU ((((((((((((....((((((((..((((...---.....))))..)))).))))........)))).))))))))---..........(((((((((....))))))...))).. ( -26.10) >DroSec_CAF1 6389 111 + 1 GAUGAAAAACAAUUUUAUCGCCGCUCCCGCUUG---UUGUUGUGGUUGUGGAUGGUUGU---UUUUGUAUUUUUAUUUUUUUUGCAGCUAUCAAAUUGGAAUUCCAAUUAAAUGGCU ((((((((((((....((((((((..((((...---.....))))..)))).))))...---..)))).))))))))........((((((..((((((....))))))..)))))) ( -32.10) >DroEre_CAF1 6186 106 + 1 GAUGAAAAACAAUUUUAUCGCUGCUCCCGCUUGGUGUUGUUGUGGUUGGGGAUUGUUUU--------UAUUUUC---UUUUUUGCAGCUACAAAAUUGGAAUUCCAAUUAAAUGGGU (((((((.....)))))))......((((.....(((.((((..(..(((((.......--------....)))---))..)..)))).))).((((((....))))))...)))). ( -25.60) >DroYak_CAF1 6578 96 + 1 GAUGAAAAACAAUUUUAUCGCUGCUCCUACUUG---UUGUUGUGAUUGUGUAUGGUUGU------------------UUUUUUGCAGCUACCAAAUUGGAAUUCCAAUUAAAUGGGU (((((((.....)))))))(((((.(((((..(---((.....)))...))).))....------------------......)))))..(((((((((....))))))...))).. ( -21.40) >consensus GAUGAAAAACAAUUUUAUCGCCGCUCCCGCUUG___UUGUUGUGGUUGUGGAUGGUUGU________UAUUUUU___UUUUUUGCAGCUACCAAAUUGGAAUUCCAAUUAAAUGGGU (((((((.....)))))))......((((............(((((((((((............................)))))))))))..((((((....))))))...)))). (-17.88 = -18.31 + 0.44)



| Location | 9,936,613 – 9,936,724 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -16.38 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.37 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9936613 111 - 22224390 ACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUGCAAA---AAUAAAAAUACAAAAAAAAAAACCAUCCACAACCACAACAA---CAAGCGGGAGCGGCGAUAAAAUUGUUUUUCAUC ...........(((((...((((((....(((((...---.((....)).......................((.(.....---...).))..)))))....))))))..))))).. ( -14.40) >DroSec_CAF1 6389 111 - 1 AGCCAUUUAAUUGGAAUUCCAAUUUGAUAGCUGCAAAAAAAAUAAAAAUACAAAA---ACAACCAUCCACAACCACAACAA---CAAGCGGGAGCGGCGAUAAAAUUGUUUUUCAUC (((.(((.((((((....)))))).))).)))...................((((---((((....((.(..((.(.....---...).))..).))........)))))))).... ( -17.00) >DroEre_CAF1 6186 106 - 1 ACCCAUUUAAUUGGAAUUCCAAUUUUGUAGCUGCAAAAAA---GAAAAUA--------AAAACAAUCCCCAACCACAACAACACCAAGCGGGAGCAGCGAUAAAAUUGUUUUUCAUC ...........(((((...(((((((((.(((((......---(......--------....)..(((((.................).))))))))).)))))))))..))))).. ( -20.23) >DroYak_CAF1 6578 96 - 1 ACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUGCAAAAAA------------------ACAACCAUACACAAUCACAACAA---CAAGUAGGAGCAGCGAUAAAAUUGUUUUUCAUC ...........(((((...((((((....(((((......------------------....((.(((.............---...))))).)))))....))))))..))))).. ( -13.89) >consensus ACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUGCAAAAAA___AAAAAUA________AAAACCAUCCACAACCACAACAA___CAAGCGGGAGCAGCGAUAAAAUUGUUUUUCAUC ...........(((((...((((((....(((((...........................................................)))))....))))))..))))).. (-12.61 = -12.37 + -0.25)

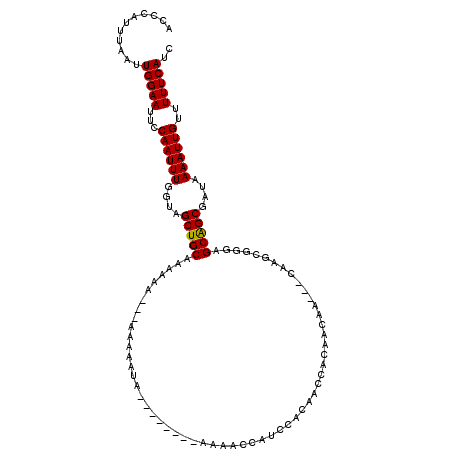

| Location | 9,936,650 – 9,936,761 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.01 |
| Mean single sequence MFE | -18.38 |
| Consensus MFE | -6.72 |
| Energy contribution | -8.24 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9936650 111 - 22224390 UGUCGCUGGGAUUUUAAUGCAAUUUUCCCAGUCCAAGACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUG---CAAA---AAUAAAAAUACAAAAAAAAAAACCAUCCACAACCACA .((((((((((..............)))))))....)))(((...((((((....)))))))))......---....---..................................... ( -18.14) >DroSec_CAF1 6426 111 - 1 UGUCGCUGGGAUUUUAAUGCAAUUUUCCCAGUCCAAGAGCCAUUUAAUUGGAAUUCCAAUUUGAUAGCUG---CAAAAAAAAUAAAAAUACAAAA---ACAACCAUCCACAACCACA (((.(((((((..............))))))).....(((.(((.((((((....)))))).))).))))---))....................---................... ( -20.44) >DroEre_CAF1 6226 103 - 1 UGUCGCGGGGAUUUUAAUGCAAUUUUCCCAGUCCAAGACCCAUUUAAUUGGAAUUCCAAUUUUGUAGCUG---CAAAAAA---GAAAAUA--------AAAACAAUCCCCAACCACA ......(((((((........((((((....(((((...........))))).......((((((....)---)))))..---)))))).--------.....)))))))....... ( -20.14) >DroYak_CAF1 6615 96 - 1 UGUCGCGGGGAUUUUAAUGCAAUUUUCCCAGUCCAAGACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUG---CAAAAAA------------------ACAACCAUACACAAUCACA (((.(((((((..((.....))..)))))..........(((...((((((....)))))))))..)).)---)).....------------------................... ( -17.80) >DroMoj_CAF1 823 98 - 1 AGUCGCUU---------GUCAAUUGCAGCAACGCAAAGCCGGUUUCAUGCCAAUUCGGCAUUCAUCGCCAAAUCCAAAGAGAUAACAAAACAAAU---A-------CAACAAACACA ....(.((---------(((..((((......))))....(((...(((((.....))))).....)))...........)))))).........---.-------........... ( -15.40) >consensus UGUCGCUGGGAUUUUAAUGCAAUUUUCCCAGUCCAAGACCCAUUUAAUUGGAAUUCCAAUUUGGUAGCUG___CAAAAAA_AUAAAAAUACAAA____AAAACCAUCCACAACCACA ....(((((((..............))))))).........((..((((((....))))))..)).................................................... ( -6.72 = -8.24 + 1.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:50 2006