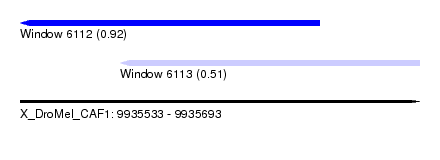
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,935,533 – 9,935,693 |
| Length | 160 |
| Max. P | 0.921498 |

| Location | 9,935,533 – 9,935,653 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -26.58 |
| Energy contribution | -25.95 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9935533 120 - 22224390 CCAACUAAAAAAUAAAACAAAAACAAAGUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCAAUUUAAUCCAUCCGGCUUUUAGUUUUGUACAUUUUCCAUC ...........................((((((((...(((((..((....((((((....)))(((((.......)))))............))).....))..))))).)))))))). ( -26.50) >DroSec_CAF1 5303 120 - 1 ACAACUAAAAAAUAAAACAAAAACAAAGUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCAAUUUAAUCCAUCCGGCUUUUAGUUUUGUACAUUUUCCAUC ...........................((((((((...(((((..((....((((((....)))(((((.......)))))............))).....))..))))).)))))))). ( -26.50) >DroEre_CAF1 5187 119 - 1 CCUACUAAAGAAUAAAACAAAAACAAAAUGGAAGA-GGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCAAUUUAAUCCAUCCGGCUUUUAGUUUUGUACAUUUUCCAUC ...........................((((((((-..(((((..((....((((((....)))(((((.......)))))............))).....))..))))).)))))))). ( -26.20) >DroYak_CAF1 5484 109 - 1 CUAACUAA-----------AAAACAAAAUGGAGGGGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCAAUUUAAUACAUCCGGCUUUUAGUUUUGUACAUUUUCCAUC ........-----------........((((((((...(((((..((....((((((....)))(((((.......)))))............))).....))..))))).)))))))). ( -25.00) >consensus CCAACUAAAAAAUAAAACAAAAACAAAAUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCAAUUUAAUCCAUCCGGCUUUUAGUUUUGUACAUUUUCCAUC ...........................((((((((...(((((..((....((((((....)))(((((.......)))))............))).....))..))))).)))))))). (-26.58 = -25.95 + -0.63)



| Location | 9,935,573 – 9,935,693 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9935573 120 - 22224390 AAAAAAAAACAUUGGGUGGGGGCUCUAGGAGCUUAGGAGCCCAACUAAAAAAUAAAACAAAAACAAAGUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCA ...........((((((..((((((...))))))....))))))...............................(((((........((((((.((((....)))))))))).))))). ( -29.50) >DroSec_CAF1 5343 116 - 1 A----AAAACAGUGGGUGGGGGGUCUCGGGGCUUAGGAGCACAACUAAAAAAUAAAACAAAAACAAAGUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCA .----..............(((((((((.(((.((.((((((..((.....((..............)).....))..)))).))...)).))).))))(((......)))...))))). ( -25.04) >DroEre_CAF1 5227 112 - 1 AAA-----A--AUGGGUAGCGAGUCUAGGGGCUUAGGACCCCUACUAAAGAAUAAAACAAAAACAAAAUGGAAGA-GGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCA ...-----.--.((((..(((((((....)))))...((((((.(((.....................))).)).-))))))......((((((.((((....))))))))))...)))) ( -26.50) >DroYak_CAF1 5524 104 - 1 AAA-----AGAGUGGGUGGUGGGUCUAGGGGCUUAGGACCCUAACUAA-----------AAAACAAAAUGGAGGGGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCA ...-----..(((((((..((((((....))))))..))))..)))..-----------.............((((..((((....((.(((((....).)))).)).))))...)))). ( -29.30) >consensus AAA_____ACAGUGGGUGGGGGGUCUAGGGGCUUAGGACCCCAACUAAAAAAUAAAACAAAAACAAAAUGGAAGAGGGGUGCAUUACAUAUGCCACGAGUGCGUUUGGGCAUAAAUCCCA ............((((......(((((((.((((((........))))..............................((((((.....))).)))....)).)))))))......)))) (-19.20 = -19.70 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:47 2006