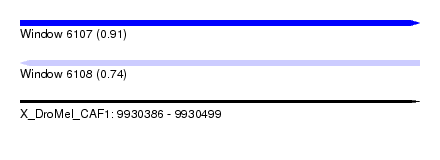
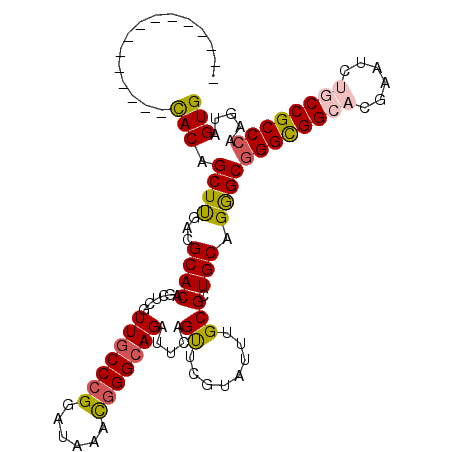
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,930,386 – 9,930,499 |
| Length | 113 |
| Max. P | 0.914590 |

| Location | 9,930,386 – 9,930,499 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -34.31 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9930386 113 + 22224390 CAUAAAUGCAGAAAUCACAGCUUGACGCACAGCUCGUUGCCCGGAUAAACGGGCAGAUUCAGUUCGUAUUUGCGCUGCAGGGCGGGCGGCACGAAUCUGCCGCCCAAGUAGUG ...............(((.((((........((((.(((((((......)))))))...((((.((....)).))))..))))((((((((......)))))))))))).))) ( -44.50) >DroVir_CAF1 2 98 + 1 ---------------UACAGCUCGAGGCACAACUCGUUACCCGGAUAGGCAGGCAGAUUCAGCUCGUACUUGCGCUGCAGAGCUGGCGGCACAAAGCGUCCGCCUAAAAAGUG ---------------...((((((((......))).............((((((((...(.....)...)))).)))).)))))(((((.((.....)))))))......... ( -30.40) >DroSec_CAF1 2 98 + 1 ---------------CACAGCUUGACGCACAGCUCGUUGCCCGGAUAAACGGGCAGAUUAAGUUCGUAUUUGCGCUGCAGGGCGGGUGGCACGAAUCUGCCGCCCAAGUAGUG ---------------(((.((((........((((.(((((((......))))))).....((.(((....)))..)).))))((((((((......)))))))))))).))) ( -41.60) >DroSim_CAF1 107 113 + 1 CAAAAAUGAAGAAAUCACAGCUUGACGCACAGCUCGUUGCCCGGAUAAACGGGCAGAUUAAGUUCGUAUUUGCGGUGCAGGGCGGGCGGCACGAAUCUGCCGCCCAAGUAGUG ...............(((.((((...((((((((..(((((((......)))))))....))))(((....))))))).))))((((((((......)))))))).....))) ( -45.20) >DroEre_CAF1 2 98 + 1 ---------------CACAGCUUGACGCACAGUUCGUUGCCCGGAUAAACGGGCAGAUUCAGUUCGUAUUUGCGCUGCAGAGCGGGCGGCACGAAUCUGCCGCCCAAGUAGUG ---------------(((.((((........((((.(((((((......)))))))...((((.((....)).))))..))))((((((((......)))))))))))).))) ( -42.90) >DroMoj_CAF1 2 98 + 1 ---------------CACAGCUCCAGGCACAUCUCAUUGCCCGGAUAUGUGGGCAGAUUCAGCUCGUACUUGCGCUGCAGGGCGGGCGGCACGAAGCGUCCGCCCAGGAAGUG ---------------((((..(((.((((........)))).)))..)))).((((.....((........)).)))).(((((((((........)))))))))........ ( -41.30) >consensus _______________CACAGCUUGACGCACAGCUCGUUGCCCGGAUAAACGGGCAGAUUCAGUUCGUAUUUGCGCUGCAGGGCGGGCGGCACGAAUCUGCCGCCCAAGUAGUG ...............(((.((((...((((......(((((((......))))))).....((........))).))).))))((((((((......)))))))).....))) (-34.31 = -34.40 + 0.09)


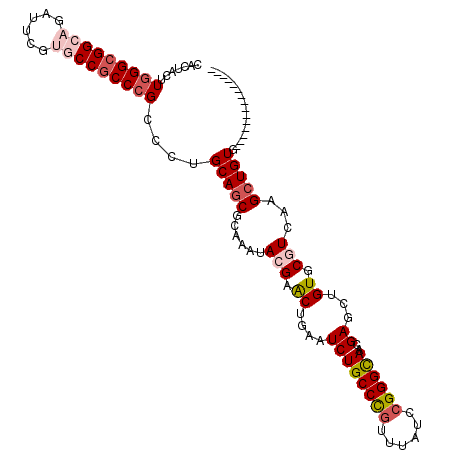
| Location | 9,930,386 – 9,930,499 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -29.22 |
| Energy contribution | -30.38 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9930386 113 - 22224390 CACUACUUGGGCGGCAGAUUCGUGCCGCCCGCCCUGCAGCGCAAAUACGAACUGAAUCUGCCCGUUUAUCCGGGCAACGAGCUGUGCGUCAAGCUGUGAUUUCUGCAUUUAUG (((..(((((((((((......)))))))(((...(((((.(......((......))((((((......))))))..).)))))))).))))..)))............... ( -40.40) >DroVir_CAF1 2 98 - 1 CACUUUUUAGGCGGACGCUUUGUGCCGCCAGCUCUGCAGCGCAAGUACGAGCUGAAUCUGCCUGCCUAUCCGGGUAACGAGUUGUGCCUCGAGCUGUA--------------- .........(((((.(((...))))))))(((((.(..((((((.(.((.((.......)).(((((....))))).))).))))))..))))))...--------------- ( -33.30) >DroSec_CAF1 2 98 - 1 CACUACUUGGGCGGCAGAUUCGUGCCACCCGCCCUGCAGCGCAAAUACGAACUUAAUCUGCCCGUUUAUCCGGGCAACGAGCUGUGCGUCAAGCUGUG--------------- (((..((((((.((((......)))).))(((...(((((.(......((......))((((((......))))))..).)))))))).))))..)))--------------- ( -33.90) >DroSim_CAF1 107 113 - 1 CACUACUUGGGCGGCAGAUUCGUGCCGCCCGCCCUGCACCGCAAAUACGAACUUAAUCUGCCCGUUUAUCCGGGCAACGAGCUGUGCGUCAAGCUGUGAUUUCUUCAUUUUUG (((..(((((((((((......)))))))).....((((.((......((......))((((((......))))))....)).))))...)))..)))............... ( -40.70) >DroEre_CAF1 2 98 - 1 CACUACUUGGGCGGCAGAUUCGUGCCGCCCGCUCUGCAGCGCAAAUACGAACUGAAUCUGCCCGUUUAUCCGGGCAACGAACUGUGCGUCAAGCUGUG--------------- (((.....((((((((......))))))))(((..((.(((((.....((......))((((((......))))))......)))))))..))).)))--------------- ( -39.20) >DroMoj_CAF1 2 98 - 1 CACUUCCUGGGCGGACGCUUCGUGCCGCCCGCCCUGCAGCGCAAGUACGAGCUGAAUCUGCCCACAUAUCCGGGCAAUGAGAUGUGCCUGGAGCUGUG--------------- ........(((((((.(((.(((((....(((......)))...))))))))....)))))))(((..((((((((........))))))))..))).--------------- ( -39.80) >consensus CACUACUUGGGCGGCAGAUUCGUGCCGCCCGCCCUGCAGCGCAAAUACGAACUGAAUCUGCCCGUUUAUCCGGGCAACGAGCUGUGCGUCAAGCUGUG_______________ .......(((((((((......)))))))))....(((((......(((.((....((((((((......))))))..))...)).)))...)))))................ (-29.22 = -30.38 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:42 2006