
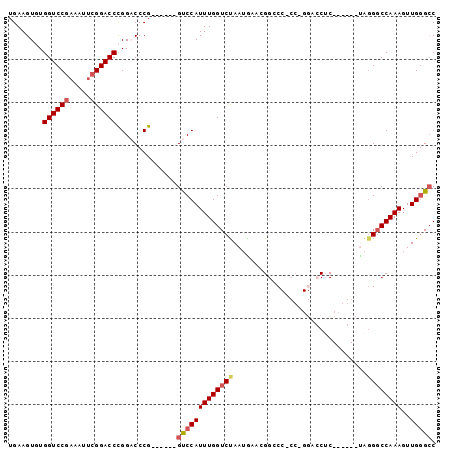
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,924,091 – 9,924,184 |
| Length | 93 |
| Max. P | 0.999665 |

| Location | 9,924,091 – 9,924,184 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9924091 93 + 22224390 UGAAGUGUGGUCCGAAAUUCGGACCCGGACCCGGUAACGGUCCAUUUGGUCUAAUGAACGGCCCGCCCGGACCUC------UAGGUCCAAAGUUGUGCC ......(.((((((.....)))))))(((((..(....(((((....((((........)))).....))))).)------..)))))........... ( -34.80) >DroSec_CAF1 21273 78 + 1 UGAAGUGUGGUCCGAAAUUCGGACCCGGACCCG------GUCCAUUUGGUCUAAUAAACGGCC---------CUC------UAGGGCCAAAGUUGGGCC ......(.((((((.....)))))))((((...------))))....((((((((....((((---------(..------..)))))...)))))))) ( -34.20) >DroSim_CAF1 21458 78 + 1 UGAAGUGUGGUCCGAAAUUCGGACCCGGACCCG------GUCCAUUUGGUCUAAUAAACGGCC---------CUC------UAGGGCCAAAGUUGGGCC ......(.((((((.....)))))))((((...------))))....((((((((....((((---------(..------..)))))...)))))))) ( -34.20) >DroEre_CAF1 21703 93 + 1 UGAAGUUUGGUCCGAAAUUCGGACCUGGUCCCG------GCCCAUUUGGUCCAAAGAAUGGGCCCCCAGGACCGCCUGGGCCGUGGCCAAAGUUGGGCC ....((((((((((.....))))))(((((.((------(((((..((((((......(((....)))))))))..))))))).))))).....)))). ( -46.90) >DroYak_CAF1 21501 93 + 1 UGAAGUUUGGUCCGAAAUGGGGACCUGGUCCCG------GUCCAUUUGGUCCAAAGAAUGGGCCACCCGGACCGCCUGGGCCAGGGCCAAAGUUGGGGC .........((((.((....((.((((((((((------((((...(((((((.....)))))))...))))))...)))))))).))....)).)))) ( -46.60) >consensus UGAAGUGUGGUCCGAAAUUCGGACCCGGACCCG______GUCCAUUUGGUCUAAUGAACGGCCC_CC_GGACCUC______UAGGGCCAAAGUUGGGCC ........((((((.....))))))..............(((((((((((((...............................))))))))..))))). (-19.86 = -20.58 + 0.72)


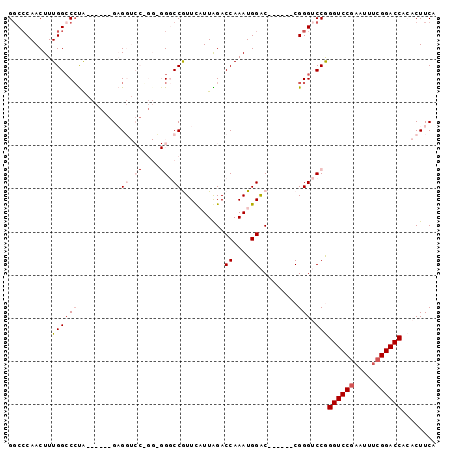
| Location | 9,924,091 – 9,924,184 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -17.40 |
| Energy contribution | -18.66 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
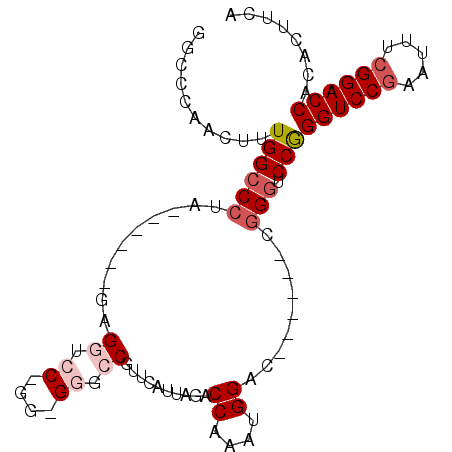
>X_DroMel_CAF1 9924091 93 - 22224390 GGCACAACUUUGGACCUA------GAGGUCCGGGCGGGCCGUUCAUUAGACCAAAUGGACCGUUACCGGGUCCGGGUCCGAAUUUCGGACCACACUUCA ........((((((((..------..))))))))(((((((((((((......)))))))((....))))))))((((((.....))))))........ ( -37.90) >DroSec_CAF1 21273 78 - 1 GGCCCAACUUUGGCCCUA------GAG---------GGCCGUUUAUUAGACCAAAUGGAC------CGGGUCCGGGUCCGAAUUUCGGACCACACUUCA (((((......(((((..------..)---------))))(((((((......)))))))------.)))))..((((((.....))))))........ ( -34.90) >DroSim_CAF1 21458 78 - 1 GGCCCAACUUUGGCCCUA------GAG---------GGCCGUUUAUUAGACCAAAUGGAC------CGGGUCCGGGUCCGAAUUUCGGACCACACUUCA (((((......(((((..------..)---------))))(((((((......)))))))------.)))))..((((((.....))))))........ ( -34.90) >DroEre_CAF1 21703 93 - 1 GGCCCAACUUUGGCCACGGCCCAGGCGGUCCUGGGGGCCCAUUCUUUGGACCAAAUGGGC------CGGGACCAGGUCCGAAUUUCGGACCAAACUUCA ((((.......)))).(((((((...(((((..((((....))))..)))))...)))))------))......((((((.....))))))........ ( -46.20) >DroYak_CAF1 21501 93 - 1 GCCCCAACUUUGGCCCUGGCCCAGGCGGUCCGGGUGGCCCAUUCUUUGGACCAAAUGGAC------CGGGACCAGGUCCCCAUUUCGGACCAAACUUCA ..(((..(((.(((....))).))).((((((..(((.(((.....))).)))..)))))------))))....(((((.......)))))........ ( -40.20) >consensus GGCCCAACUUUGGCCCUA______GAGGUCC_GG_GGGCCGUUCAUUAGACCAAAUGGAC______CGGGUCCGGGUCCGAAUUUCGGACCACACUUCA ..........((((((..........((.((....)).))..........((....)).........))).)))((((((.....))))))........ (-17.40 = -18.66 + 1.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:40 2006