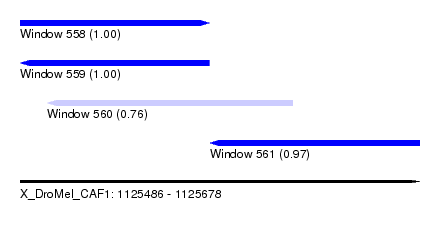
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,125,486 – 1,125,678 |
| Length | 192 |
| Max. P | 0.999721 |

| Location | 1,125,486 – 1,125,577 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
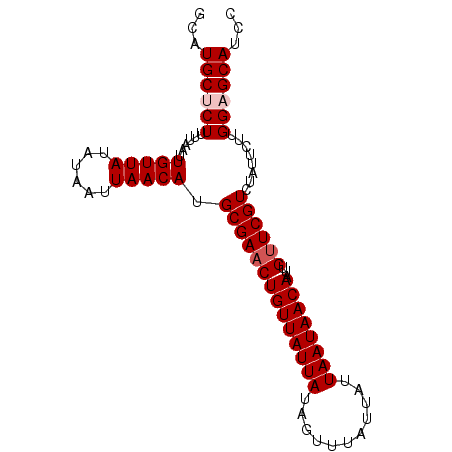
>X_DroMel_CAF1 1125486 91 + 22224390 AUAUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGUUCGUCUAUUCUUGGAGCAUCC ..(((((((......(((((......))))).((((((((((((((...........))))))))...))))))........))))))).. ( -21.50) >DroSec_CAF1 22213 91 + 1 GCAUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGUUCGUCUAUUCUUGGAGCAUCC ..(((((((......(((((......))))).((((((((((((((...........))))))))...))))))........))))))).. ( -21.60) >DroSim_CAF1 23563 91 + 1 GCAUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGUUCGUCUAUUCUUGGAGCAUCC ..(((((((......(((((......))))).((((((((((((((...........))))))))...))))))........))))))).. ( -21.60) >DroEre_CAF1 23377 91 + 1 GCCUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGUUCGUCUAUUCUUGGCGCAUCC (((............(((((......))))).((((((((((((((...........))))))))...))))))........)))...... ( -18.30) >DroYak_CAF1 22955 91 + 1 GCCUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGCUCGUCUAUUCUUGGCGCAUCC (((............(((((......))))).((((..((((((((...........)))))))).))))............)))...... ( -15.40) >consensus GCAUGCUCUUUUAAUUGUUAUAUAAUUAACAUGCGAACUGUUAUUAUAGUUUAUUAUUAAUAACAAUUGUUCGUCUAUUCUUGGAGCAUCC ...((((((......(((((......))))).((((((((((((((...........))))))))...))))))........))))))... (-17.90 = -18.50 + 0.60)

| Location | 1,125,486 – 1,125,577 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
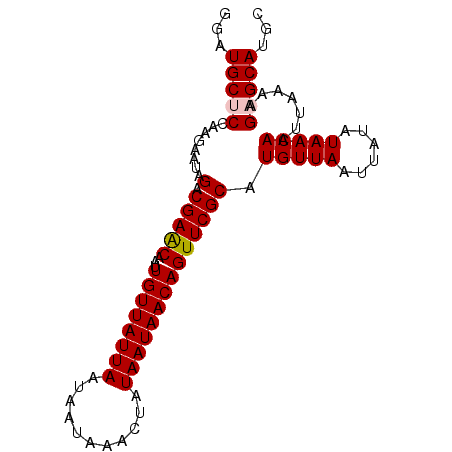
>X_DroMel_CAF1 1125486 91 - 22224390 GGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAUAU ..((((((........(.(((((...((((((((...........)))))))))))))).(((((......))))).......)))))).. ( -19.30) >DroSec_CAF1 22213 91 - 1 GGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAUGC (.((((((........(.(((((...((((((((...........)))))))))))))).(((((......))))).......)))))).) ( -19.70) >DroSim_CAF1 23563 91 - 1 GGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAUGC (.((((((........(.(((((...((((((((...........)))))))))))))).(((((......))))).......)))))).) ( -19.70) >DroEre_CAF1 23377 91 - 1 GGAUGCGCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAGGC ......(((.......(.(((((...((((((((...........)))))))))))))).(((((......)))))............))) ( -15.50) >DroYak_CAF1 22955 91 - 1 GGAUGCGCCAAGAAUAGACGAGCAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAGGC ......(((............(((((((((((((...........))))))))))).)).(((((......)))))............))) ( -16.30) >consensus GGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAUUAAAAGAGCAUGC ...(((((........(.(((((...((((((((...........)))))))))))))).(((((......))))).......)))))... (-16.04 = -16.28 + 0.24)

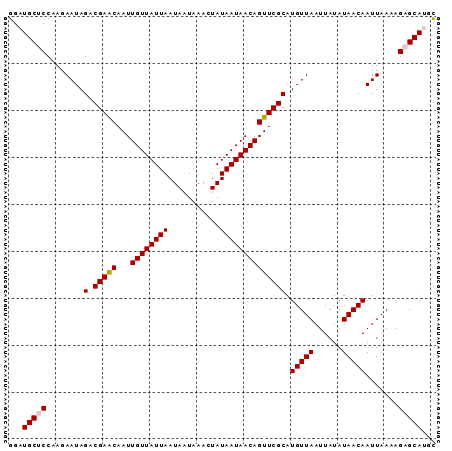
| Location | 1,125,499 – 1,125,617 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -16.88 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1125499 118 - 22224390 UUGCCAAAAAAAAAGCACUGAAACGAAAUCAAAAACCGUCGGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU .(((.........((((((((..((...........)))))).))))............((((...((((((((...........)))))))))))))))(((((......))))).. ( -17.40) >DroSec_CAF1 22226 116 - 1 UUGCCAAAA--AAAGCACUGAAACGAAAUCAAAAACCGUCGGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU .(((.....--..((((((((..((...........)))))).))))............((((...((((((((...........)))))))))))))))(((((......))))).. ( -17.40) >DroSim_CAF1 23576 117 - 1 UUGCCAAAAA-AAAACACUGAAACGAAAUCAAAAACCGUCGGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU .(((......-.........................((((.....(.....)....))))...(((((((((((...........))))))))))).)))(((((......))))).. ( -14.40) >DroEre_CAF1 23390 115 - 1 UUGCCAAA---AAAGCACUGAAUCGAAAUCAAAAACCGUCGGAUGCGCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU .(((....---...(((((((..((...........)))))).))).............((((...((((((((...........)))))))))))))))(((((......))))).. ( -17.30) >DroYak_CAF1 22968 115 - 1 UUGCCAAA---AAAGCAAUGAAACGAAAUGAAAAACCGUCGGAUGCGCCAAGAAUAGACGAGCAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU ((((....---...))))..................((((.....(.....)....)))).(((((((((((((...........))))))))))).)).(((((......))))).. ( -17.90) >consensus UUGCCAAAA__AAAGCACUGAAACGAAAUCAAAAACCGUCGGAUGCUCCAAGAAUAGACGAACAAUUGUUAUUAAUAAUAAACUAUAAUAACAGUUCGCAUGUUAAUUAUAUAACAAU .(((..........(((((((..((...........)))))).))).............((((...((((((((...........)))))))))))))))(((((......))))).. (-14.84 = -15.08 + 0.24)

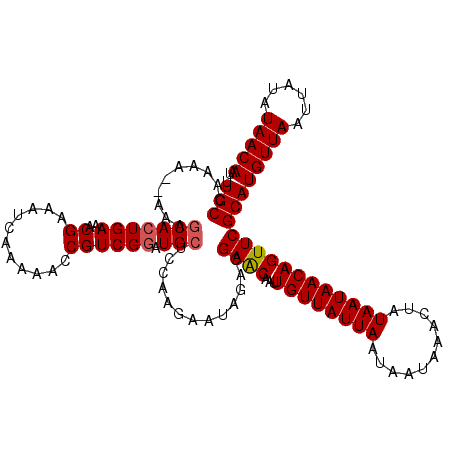
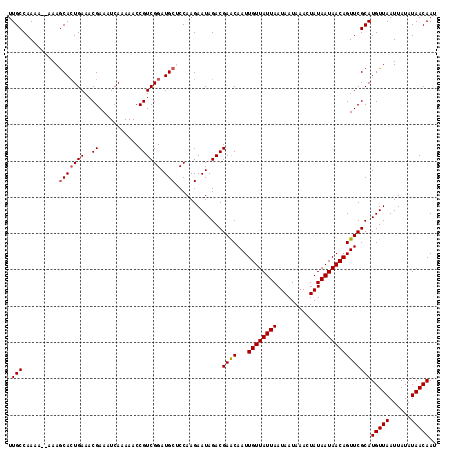
| Location | 1,125,577 – 1,125,678 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1125577 101 - 22224390 UCGGUGUUACGCUUUUUGAUCCAUUAAAUCAGACGGCAAACGCAGCACACAGC-----------AAAAUA--------UAUUGCCAAAAAAAAAGCACUGAAACGAAAUCAAAAACCGUC ((((((((..(((.((((((.......)))))).)))..............((-----------((....--------..)))).........))))))))................... ( -19.40) >DroSec_CAF1 22304 99 - 1 UCGGUGUUAUGCUUUUUGAUCCAUUAAAUCAGACGGCAAACGCAGCACACAAC-----------GAAAUA--------UAUUGCCAAAA--AAAGCACUGAAACGAAAUCAAAAACCGUC ((((((((.((((.((((((.......)))))).))))...((((........-----------......--------..)))).....--..))))))))................... ( -18.09) >DroSim_CAF1 23654 100 - 1 UCGGUGUUACGCUUUUUGAUCCAUUAAAUCAGACGGCAAACGCAGCACACAAC-----------GAAAUA--------UAUUGCCAAAAA-AAAACACUGAAACGAAAUCAAAAACCGUC ((((((((..(((.((((((.......)))))).)))....((((........-----------......--------..))))......-..))))))))................... ( -17.39) >DroEre_CAF1 23468 100 - 1 UCGGUGUUACGCUUUUUGAUCCAUAAAAUCAGACGGCAAACGCAACACACAAA-----------AAAAUA------UAUAUUGCCAAA---AAAGCACUGAAUCGAAAUCAAAAACCGUC ((((((((..(((.((((((.......)))))).)))....((((........-----------......------....))))....---..))))))))................... ( -17.57) >DroYak_CAF1 23046 117 - 1 UCGGUGUUACGCUUUUUGAUCCAUAAAAUCAGACGGCAAACGCAACACACACAACAACAACAACAAAAUAUAUAUGUAUAUUGCCAAA---AAAGCAAUGAAACGAAAUGAAAAACCGUC ((((((((..(((.((((((.......)))))).))).)))))...................................((((((....---...))))))...))).............. ( -17.80) >consensus UCGGUGUUACGCUUUUUGAUCCAUUAAAUCAGACGGCAAACGCAGCACACAAC___________AAAAUA________UAUUGCCAAAA__AAAGCACUGAAACGAAAUCAAAAACCGUC ((((((((..(((.((((((.......)))))).)))....((((...................................)))).........))))))))................... (-15.95 = -15.75 + -0.20)

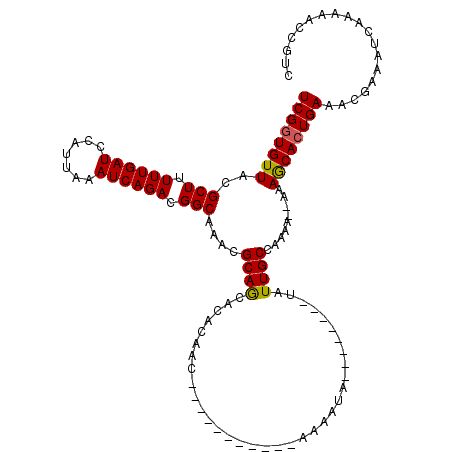
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:51 2006