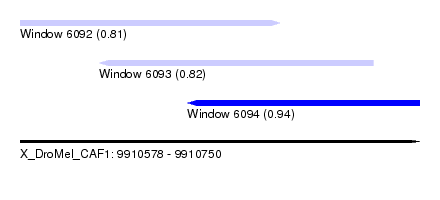
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,910,578 – 9,910,750 |
| Length | 172 |
| Max. P | 0.938516 |

| Location | 9,910,578 – 9,910,690 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
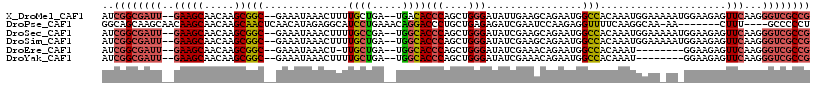
>X_DroMel_CAF1 9910578 112 + 22224390 AUCGGCGAUU--GAAGCAACAAGCGGC--GAAAUAAACUUUUGCUGA--UGACACCCAGCUGGGAUAUUGAAGCAGAAUGGCCACAAAUGGAAAAAUGGAAGAGUUCAAGGGUCGCCG ..((((((((--...((..(((.((((--((((.....)))))))).--.....(((....)))...)))..)).((((..(((............)))....))))...)))))))) ( -31.40) >DroPse_CAF1 10207 106 + 1 GGCAGCAAGCAACAAGCAACAAGCAACUCAACAUAGAGGCAUCCUGAAACAGGACCCUGCUGAGAGAUCGAAUCCAAGAGGUUUUCAAGGCAA-AA-------CUUU----GCCCCCU (((((...((.....((.....))..(((......))))).(((((...)))))..)))))(((((.((........))..)))))..(((((-..-------..))----))).... ( -25.60) >DroSec_CAF1 7984 112 + 1 AUCGGCGAUU--GAAGCAACAAGCGGC--GAAAUAAACUUUUGCCGA--UGGCACCCAGCUGGGAUAUCGAAGCAGAAUGGCCACAAAUGGAAAAAUGGAAGAGUUCAAGGGUCGCCG ..((((((..--...((......((((--((((.....)))))))).--..)).(((.(((..(....)..))).((((..(((............)))....))))..))))))))) ( -33.70) >DroSim_CAF1 8028 112 + 1 AUCGGCGAUU--GAAGCAACAAGCGGC--GAAAUAAACUUUUGCUGA--UGGCACCCAGCUGGGAUAUCGAAGCAGAAUGGCCACAAAUGGAAAAAUGGAAGAGUUCAAGGGUCGCCG ..((((((..--...((......((((--((((.....)))))))).--..)).(((.(((..(....)..))).((((..(((............)))....))))..))))))))) ( -31.50) >DroEre_CAF1 8296 103 + 1 AUCGGCGAUU--GAAGCAACAAGCGGC--GAAAUAAACU-UUGCUGA--UGGCACCCAGCUGGGAUAUCGAAACAGAAUGGCCACAAAU--------GGAAGAGUUCAAGGGUCGCCG ..((((((((--((.((......((((--(((......)-)))))).--..)).(((....)))...))......((((..(((....)--------))....))))...)))))))) ( -30.60) >DroYak_CAF1 7927 104 + 1 AUCGGCGAUU--GAAGCAACAAGCGGC--GAAAUAAACUUUUGCUGA--UGGCACCCAGCUGGGAUAUCGAAACAGAAUGGCCACAAAU--------GGAAGAGUUCAAGGGUCGCCG ..((((((((--((.((......((((--((((.....)))))))).--..)).(((....)))...))......((((..(((....)--------))....))))...)))))))) ( -31.10) >consensus AUCGGCGAUU__GAAGCAACAAGCGGC__GAAAUAAACUUUUGCUGA__UGGCACCCAGCUGGGAUAUCGAAGCAGAAUGGCCACAAAUGGAA_AA_GGAAGAGUUCAAGGGUCGCCG ..((((((((..(((((.....))(((..............((((.....))))(((....)))................))).....................)))...)))))))) (-19.06 = -19.62 + 0.56)


| Location | 9,910,612 – 9,910,730 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.34 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.14 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
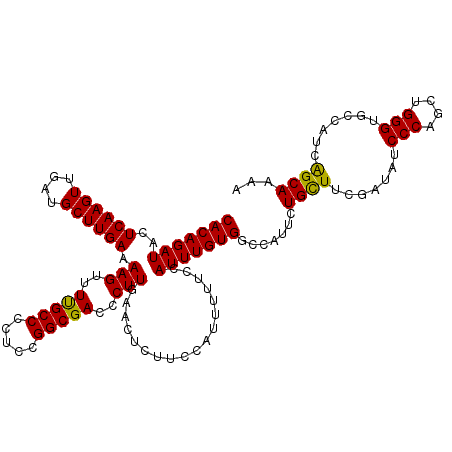
>X_DroMel_CAF1 9910612 118 - 22224390 CACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCAUUCUGCUUCAAUAUCCCAGCUGGGUGUCAUCAGCAAAA (((((((..((((((....)))))).(((..(((((.....)))))..)))..................))))))).......((((.......(((....))).......))))... ( -27.74) >DroSec_CAF1 8018 118 - 1 CACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCAUUCUGCUUCGAUAUCCCAGCUGGGUGCCAUCGGCAAAA (((((((..((((((....)))))).(((..(((((.....)))))..)))..................))))))).......(((..((((..(((....)))....)))))))... ( -28.60) >DroSim_CAF1 8062 118 - 1 CACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCAUUCUGCUUCGAUAUCCCAGCUGGGUGCCAUCAGCAAAA (((((((..((((((....)))))).(((..(((((.....)))))..)))..................))))))).......((((..(((..(((....)))....)))))))... ( -27.80) >DroEre_CAF1 8330 109 - 1 CACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCC--------AUUUGUGGCCAUUCUGUUUCGAUAUCCCAGCUGGGUGCCAUCAGCAA-A (((((((..((((((....)))))).(((..(((((.....)))))..)))..........--------)))))))......................(((((......)))))..-. ( -26.30) >DroYak_CAF1 7961 110 - 1 CACAGAUAAUCAAGUUGAUGCUUGAAAAGUUUCGCCCCUCCGGCGACCCUUGAACUCUUCC--------AUUUGUGGCCAUUCUGUUUCGAUAUCCCAGCUGGGUGCCAUCAGCAAAA (((((((..((((((....)))))).(((..(((((.....)))))..)))..........--------)))))))......................(((((......))))).... ( -28.60) >consensus CACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCAUUCUGCUUCGAUAUCCCAGCUGGGUGCCAUCAGCAAAA (((((((..((((((....)))))).(((..(((((.....)))))..)))..................))))))).......((((.......(((....))).......))))... (-27.68 = -27.12 + -0.56)


| Location | 9,910,650 – 9,910,750 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.95 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
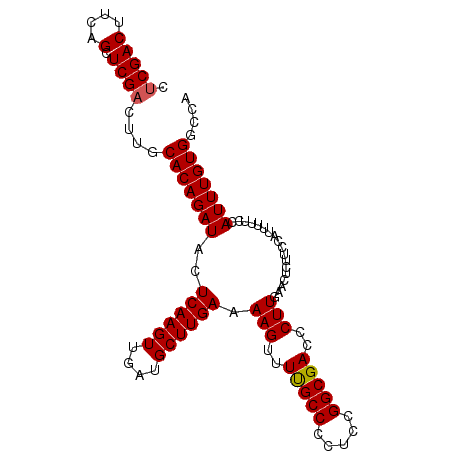
>X_DroMel_CAF1 9910650 100 - 22224390 CUCGACUUCAGCUCGACUUGCACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCA ...((.((((((((((((((..........)))))))).)))....(((..(((((.....)))))..)))))).))..((((..........))))... ( -21.90) >DroSec_CAF1 8056 100 - 1 CUCGACUUCAGCUCGACUUGCACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCA ...((.((((((((((((((..........)))))))).)))....(((..(((((.....)))))..)))))).))..((((..........))))... ( -21.90) >DroSim_CAF1 8100 100 - 1 CUCGACUUCAGCUCGACUUGCACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCA ...((.((((((((((((((..........)))))))).)))....(((..(((((.....)))))..)))))).))..((((..........))))... ( -21.90) >DroEre_CAF1 8367 92 - 1 CGCGACUUCAGCUCGACUUGCACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCC--------AUUUGUGGCCA .((.((((((((((((((((..........)))))))).)).))))(((..(((((.....)))))..)))..........--------....)).)).. ( -24.10) >DroYak_CAF1 7999 92 - 1 CUCGACUUCAGCUCGACUUGCACAGAUAAUCAAGUUGAUGCUUGAAAAGUUUCGCCCCUCCGGCGACCCUUGAACUCUUCC--------AUUUGUGGCCA .(((((....).))))....(((((((..((((((....)))))).(((..(((((.....)))))..)))..........--------))))))).... ( -24.10) >consensus CUCGACUUCAGCUCGACUUGCACAGAUACUCAAGUUGAUGCUUGAAAAGUUUUGCCCCUCCGGCGACCCUUGAACUCUUCCAUUUUUCCAUUUGUGGCCA .(((((....).))))....(((((((..((((((....)))))).(((..(((((.....)))))..)))..................))))))).... (-21.50 = -21.54 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:30 2006