
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,889,016 – 9,889,315 |
| Length | 299 |
| Max. P | 0.962376 |

| Location | 9,889,016 – 9,889,128 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889016 112 + 22224390 UCAGACAAUCAAUGGUGCUAUUAGUGUGAAAACUAAUCCAAUUAUACAAACGACAACCAACAGGCCAAAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAA ........((((((.....((((((......)))))).........................(((((..((.....))..)))))(((((........)))))..)))))). ( -20.60) >DroSec_CAF1 20293 112 + 1 UCAGACAAUCAAUGGUGCUAUUAGUGUAAACACUAAUCCAGUUAUACAAACAACAACCAACAGGCCAGAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAA ........((((((((...(((((((....)))))))...(((.....)))....))).....(((.(((....))).))).((((((((........))))))))))))). ( -24.90) >DroEre_CAF1 28949 112 + 1 UCAGACAAUCAAUGGUGCUAUUAGUGUGAACGAUAAUCCAGUUAUACAAGCCACAACCAACACGCCAGAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAA ........(((((((((((....((((((..(......)..)))))).)))....)))....((((.(((....))).))))((((((((........))))))))))))). ( -25.20) >DroYak_CAF1 12819 112 + 1 UCAGGCAAUCAAUGGUGCUAUUGGUGUGAACAUUGAUUCGGUUAUACAAACAACAACCACCAGGCCAAAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAA ...(((......((((...((..(((....)))..))..((((...........)))))))).))).........((((...((((((((........)))))))).)))). ( -28.50) >consensus UCAGACAAUCAAUGGUGCUAUUAGUGUGAACACUAAUCCAGUUAUACAAACAACAACCAACAGGCCAAAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAA ........((((((.((..(((((((....))))))).)).......................((((..((.....))..))))((((((........)))))).)))))). (-19.50 = -19.88 + 0.38)



| Location | 9,889,054 – 9,889,165 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -10.32 |
| Energy contribution | -11.92 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889054 111 + 22224390 CAAUUAUACAAACGACAACCAACAGGCCAAAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC---CAGAGCUUAACACACAAACAACAA ........................((((...((.....))....((((((((........))))))))........))))..(((((---....)))))............... ( -21.70) >DroSec_CAF1 20331 111 + 1 CAGUUAUACAAACAACAACCAACAGGCCAGAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC---CUGAGCUUAACACACAAACAACAA ......................((((((((.((.((.((((...((((((((........)))))))).))))..))..))))).))---)))..................... ( -27.10) >DroEre_CAF1 28987 114 + 1 CAGUUAUACAAGCCACAACCAACACGCCAGAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGCUUACACAGCUUAACACACAGACAACAA ..(((......(((..........((((.(((....))).))))((((((((........))))))))........)))...((((((.....)))))).......)))..... ( -27.30) >DroYak_CAF1 12857 111 + 1 CGGUUAUACAAACAACAACCACCAGGCCAAAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACAGAGC---CACAGCUUAACACACAAACCACAA .((((...................((((...((.....))....((((((((........))))))))........))))...((((---....))))........)))).... ( -25.10) >DroAna_CAF1 88497 101 + 1 ----CACCCAAACAACAAUCGACACAUUAGAUAAAGAAGAAGCCGUUGAACAAACCACAAACUUGGUCAUUGAGACAACUAGUGA-------CAGCAUACUAGAU--ACACCGA ----..............(((.......................(((......((((......))))......)))..((((((.-------.....))))))..--....))) ( -11.00) >consensus CAGUUAUACAAACAACAACCAACAGGCCAGAUGACGAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC___CACAGCUUAACACACAAACAACAA ........................((((...((.....))....((((((((........))))))))........))))..(((((.......)))))............... (-10.32 = -11.92 + 1.60)

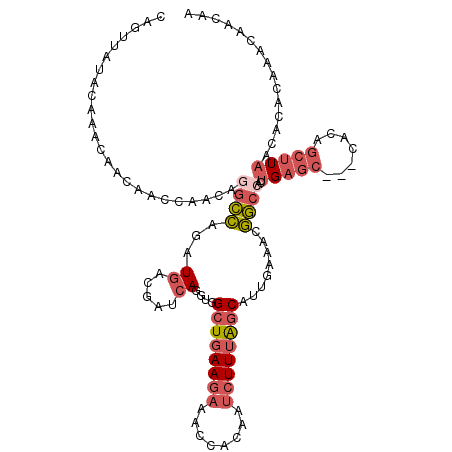

| Location | 9,889,089 – 9,889,205 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889089 116 + 22224390 GAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC---CAGAGCUUAACACACAAACAACAAUUCCCAAAACAGAAGCCAAUAGCGAAACAGUAACAGCCAU ..((((...((((((((........)))))))).))))...(((.(((((((---....)))...............................((.....))....))))....))).. ( -22.10) >DroSec_CAF1 20366 116 + 1 GAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC---CUGAGCUUAACACACAAACAACAACUUCCAAAACAGAAGCCAAUAGCGAAACACUAACAGCCAU ..((((...((((((((........)))))))).))))...(((...(((((---....)))))...............((((.......))))....................))).. ( -22.50) >DroEre_CAF1 29022 119 + 1 GAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGCUUACACAGCUUAACACACAGACAACAACUCCCAAAACAGAAGCCAACAGUGAAACACUAACAGCCAU ..((((...((((((((........)))))))).))))...(((...((((((.....))))))...................................((((...))))....))).. ( -23.50) >consensus GAUCAGGUGGCUGAAGAAACCACAAUCUUUAGCAUUGAAACGGCCACUGAGC___CAGAGCUUAACACACAAACAACAACUCCCAAAACAGAAGCCAAUAGCGAAACACUAACAGCCAU ..((((...((((((((........)))))))).))))...(((...(((((.......))))).............................((.....))............))).. (-20.52 = -20.30 + -0.22)

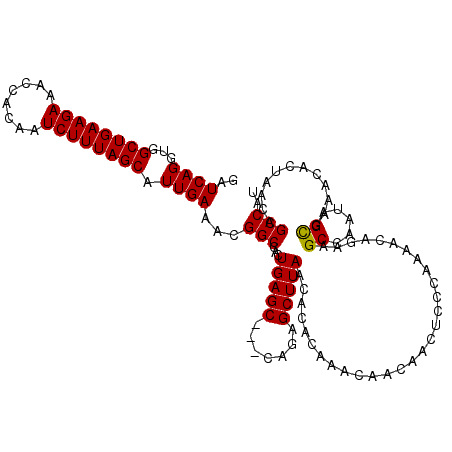

| Location | 9,889,128 – 9,889,241 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.07 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889128 113 + 22224390 ACGGCCACUGAGC---CAGAGCUUAACACACAAACAACAAUUCCCAAAACAGAAGCCAAUAGCGAAACAGUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCU ..(((((((((.(---(((.((((............................)))))...........(((((((((...(((....)))))))))))).))).)))))..)))). ( -30.29) >DroSec_CAF1 20405 113 + 1 ACGGCCACUGAGC---CUGAGCUUAACACACAAACAACAACUUCCAAAACAGAAGCCAAUAGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCU ..(((((((((.(---(((((((.................((((.......)))).....))).......(((((((...(((....))))))))))))).)).)))))..)))). ( -27.25) >DroEre_CAF1 29061 116 + 1 ACGGCCACUGAGCUUACACAGCUUAACACACAGACAACAACUCCCAAAACAGAAGCCAACAGUGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGCUCAGUUUGGUCU ..((((..((((((.....)))))).....................((((.((.((((..((((...))))((((((...(((....)))))))))....)))))).)))))))). ( -30.20) >DroYak_CAF1 12931 113 + 1 ACGGCCACAGAGC---CACAGCUUAACACACAAACCACAACACCCAAAACAGAAGCCAACGGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGCCA ..(((((..((((---....)))).................(((((........(((...))).......(((((((...(((....))))))))))...))))).....))))). ( -32.60) >consensus ACGGCCACUGAGC___CACAGCUUAACACACAAACAACAACUCCCAAAACAGAAGCCAACAGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCU ..((((..(((((.......))))).....................((((....((.....))((..((.(((((((...(((....))))))))))...))..)).)))))))). (-25.20 = -25.07 + -0.13)

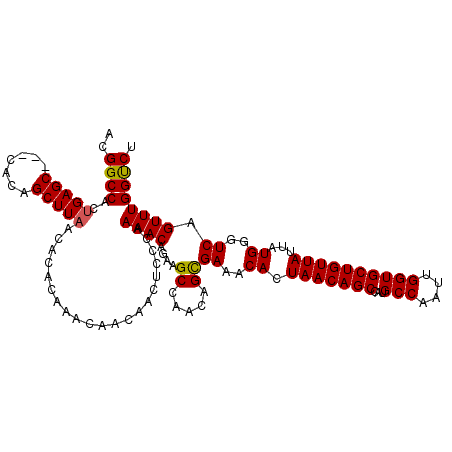

| Location | 9,889,165 – 9,889,278 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889165 113 + 22224390 UUCCCAAAACAGAAGCCAAUAGCGAAACAGUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCUAAACACACAAUCAA---CAACCGCUGUUGACAAUGACAAC ..............(.((((((((....(((((((((...(((....)))))))))))).(((..(.....)..))).............---....)))))))).)......... ( -28.40) >DroSec_CAF1 20442 113 + 1 CUUCCAAAACAGAAGCCAAUAGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCUAAACACACAAUCAA---CAACCAAUGCAGACAAUGACAUC ...(((........((.....)).......(((((((...(((....))))))))))...)))(((.(((((((................---..))))).)).)))......... ( -20.07) >DroEre_CAF1 29101 113 + 1 CUCCCAAAACAGAAGCCAACAGUGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGCUCAGUUUGGUCUAAACACACAAUCAA---CAACCACUGUAGACAAUGACAUC ...........((.((((..((((...))))((((((...(((....)))))))))....)))))).(((..(((((.............---.........)))))...)))... ( -23.15) >DroYak_CAF1 12968 116 + 1 CACCCAAAACAGAAGCCAACGGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGCCAAAACACACAGUCAACAACAACCACUGUGGACAAUGACAAC ...(((........(((...))).......(((((((...(((....))))))))))...)))((((((((.....)))).((((((...........)))))).....))))... ( -28.50) >consensus CUCCCAAAACAGAAGCCAACAGCGAAACACUAACAGCCAUGCCAAUUGGUGCUGUUAUUAUGGGUCAGUUUGGUCUAAACACACAAUCAA___CAACCACUGUAGACAAUGACAAC ...(((((......((.....))((..((.(((((((...(((....))))))))))...))..))..)))))........................((.((....)).))..... (-18.42 = -18.05 + -0.37)



| Location | 9,889,205 – 9,889,315 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9889205 110 - 22224390 CACUGCUAAUUGUGCUUGUUGUUUGAGCAUUUAGUUGGUUGUCAUUGUCAACAGCGGUUG---UUGAUUGUGUGUUUAGACCAAACUGACCCAUAAUAACAGCACCAAUUGGC ....((((((((((((.((((((.......((((((((((..(((.((((((((...)))---)))))...)))....)))).)))))).....)))))))))).)))))))) ( -34.80) >DroSec_CAF1 20482 110 - 1 CACUGCUAAUUGUGCUUGUUGUUUGAGCAUUUAGUUGGAUGUCAUUGUCUGCAUUGGUUG---UUGAUUGUGUGUUUAGACCAAACUGACCCAUAAUAACAGCACCAAUUGGC ....((((((((((((.(((((((((((((.((((..(...(((.((....)).)))...---)..)))).))))))))))(.....).........))))))).)))))))) ( -29.10) >DroEre_CAF1 29141 110 - 1 CACUGCUAAUUGUGCUUGUUGUUUGAGUAUUUAGUUGGAUGUCAUUGUCUACAGUGGUUG---UUGAUUGUGUGUUUAGACCAAACUGAGCCAUAAUAACAGCACCAAUUGGC ....((((((((((((((.....)))))))......((....(((((....)))))((((---(((.(((((.((((((......)))))))))))))))))).))))))))) ( -31.20) >DroYak_CAF1 13008 113 - 1 CACUGCUAAUUGAGCUUGUUGUUUGAGUAUUUAGUUGGUUGUCAUUGUCCACAGUGGUUGUUGUUGACUGUGUGUUUUGGCCAAACUGACCCAUAAUAACAGCACCAAUUGGC ....((((((((.(..(((((((....(((...((..(((((((..(..((((((.(.......).))))))..)..))))..)))..))..)))..)))))))))))))))) ( -32.50) >consensus CACUGCUAAUUGUGCUUGUUGUUUGAGCAUUUAGUUGGAUGUCAUUGUCUACAGUGGUUG___UUGAUUGUGUGUUUAGACCAAACUGACCCAUAAUAACAGCACCAAUUGGC ....((((((((((((.((((((.......((((((....(((......((((.(((((......)))))))))....)))..)))))).....)))))))))).)))))))) (-24.48 = -24.10 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:21 2006