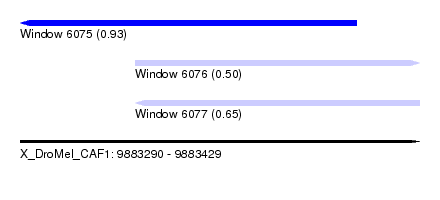
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,883,290 – 9,883,429 |
| Length | 139 |
| Max. P | 0.926299 |

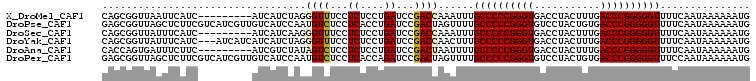
| Location | 9,883,290 – 9,883,407 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9883290 117 - 22224390 UAGGGUUUCCUCUCCUGAUCCGACCAAAUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUGACCGAGCUCGGCAUUGAUUUCCUUGGCCUGCGCCAUAGCC .((((..((......................((((((((((.(.......).))))))))))...........((((.(((....)))))))))..))))((((....))))..... ( -36.90) >DroVir_CAF1 14470 117 - 1 AAAUGUUUCCUCAGCAUUUUCCGUCAAGUUAGCCCCCGGGUGGCCUAAACUGACCCGGGGGUUUUCAAUAAAAAAUGACUCAGUUCAGAAUUGAUUUCGCGUGCCUGCUCCAUAGCC .............((((.....((((((..(((((((((((((......)).)))))))))))..).........(((......)))...))))).....))))..(((....))). ( -31.50) >DroGri_CAF1 25253 107 - 1 ----------UCAGGUGAUCCGAUCAAUUUUGCCCCCGGGUGACCUACGCUGACCCGGGGGUUUUCAAUAAAAAAUGACUUAAUUCGGAAUUGAUUUCACGUGCCUGUUCCAUAGCC ----------.((((((....((((((((((((((((((((.(.......).))))))))))..(((........)))........)))))))))).....)))))).......... ( -35.50) >DroMoj_CAF1 32446 117 - 1 AAUUGCAUUCUCAGCCUGCUCCUCUACUUUUGCCCCCGGGUGACCUACACUGACCCGGGGGUUUUCAAUAAAAAAUGACUCAAUUCAGAGUUGAUUUCGCGUGCCUGCUCCAUCGCU .............((.(((............((((((((((.(.......).))))))))))..............(((((......)))))......))).))..((......)). ( -31.50) >DroAna_CAF1 83152 117 - 1 UAUAGUCUCCUCUCCUGAUCCGACUAAUUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUGACCGAGCUCGGCAUUAAUUUCUCUAGCCUGCUCCAUUGCC ..(((((...((....))...))))).....((((((((((.(.......).))))))))))...........((((...((((..(((.............))).))))))))... ( -35.12) >DroPer_CAF1 16093 117 - 1 CAAUGUCUCCUCACCAGAUCCGACUAGUUUUGCCCCCGGGUGUCCUACUGUGACCCGGGGGUUUCCAAUAAAAAAUGACCGAGUUCGGCAUUGAUCUCCUUGGCCUGCUCCAUGGCU ...............(((((...........((((((((((...........))))))))))...........((((.(((....))))))))))))....((((........)))) ( -36.20) >consensus AAAUGUCUCCUCACCUGAUCCGACCAAUUUUGCCCCCGGGUGACCUACUCUGACCCGGGGGUUUUCAAUAAAAAAUGACCGAGUUCGGAAUUGAUUUCCCGUGCCUGCUCCAUAGCC ...............................((((((((((.(.......).))))))))))................((......))..................((......)). (-21.59 = -21.03 + -0.55)



| Location | 9,883,330 – 9,883,429 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9883330 99 + 22224390 CAUUUUUUAUUGAAAACCCCCGGGUCAAAGUAGGUCACCCGGGGGCAAAUUUGGUCGGAUCAGGAGAGGAAACCCUAGAUGAU---------GAUGAAUUAACCGCUG ................(((((((((...........))))))))).....((.((((.((((((...(....)))).)))..)---------))).)).......... ( -27.10) >DroPse_CAF1 11015 108 + 1 CAUUUUUUAUUGAAAACCCCCGGGUCACAGUAGGACACCCGGGGGCAAAACUAGUCGGAUCAGGUGAGGAGACAUUGGAUGACAACGAUGACGAAGAGCUAACCGCUC ..(((.((((((....(((((((((..(....)...)))))))))........((((..((((....(....).)))).))))..)))))).)))((((.....)))) ( -33.90) >DroSec_CAF1 14679 99 + 1 CAUUUUUUAUUGAAAACCCCCGGGUCAAAGUAGGUCACCCGGGGGCAAAUUUGGUCGGAUCAGGAGAGGAAACCCUUGAUGAU---------GAUGAAAUAACCGCUG ................(((((((((...........)))))))))...((((.((((.((((((.(.(....))))))))..)---------))).))))........ ( -32.60) >DroYak_CAF1 7044 105 + 1 CAUUUUUUAUUGAAAACCCCCGGGUCAAAGUAGGUCACCCGGGGGCAAAGUUGGUCGGAUCAGGAGAGGAAACCCUAGAUGAUGAUGAU---GAUGAAAUAACCGCUG ....(((((((.....(((((((((...........)))))))))........((((.((((..((.(....).))...))))..))))---)))))))......... ( -30.60) >DroAna_CAF1 83192 99 + 1 CAUUUUUUAUUGAAAACCCCCGGGUCAAAGUAGGUCACCCGGGGGCAAAAUUAGUCGGAUCAGGAGAGGAGACUAUAGACGAU---------GAAGAAAUCACUGGUG ..((((((((((....(((((((((...........)))))))))......(((((...((....))...)))))....))))---------)))))).......... ( -32.30) >DroPer_CAF1 16133 108 + 1 CAUUUUUUAUUGGAAACCCCCGGGUCACAGUAGGACACCCGGGGGCAAAACUAGUCGGAUCUGGUGAGGAGACAUUGGAUGACAACGAUGACGAAGAGCUAACCGCUC ..(((.((((((....(((((((((..(....)...)))))))))........(((..(((..(((......)))..))))))..)))))).)))((((.....)))) ( -37.00) >consensus CAUUUUUUAUUGAAAACCCCCGGGUCAAAGUAGGUCACCCGGGGGCAAAAUUAGUCGGAUCAGGAGAGGAAACCCUAGAUGAU_________GAAGAAAUAACCGCUG ................(((((((((...........))))))))).......((.(((.((....))(....).............................))))). (-22.54 = -22.32 + -0.22)



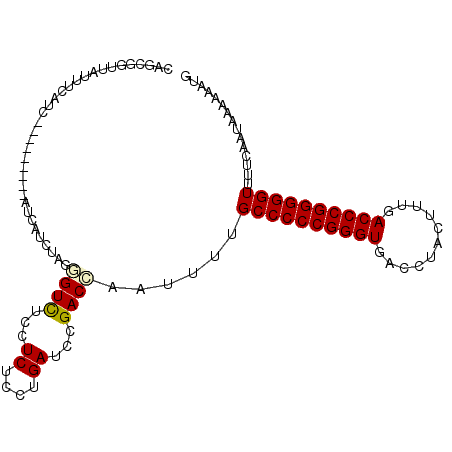
| Location | 9,883,330 – 9,883,429 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9883330 99 - 22224390 CAGCGGUUAAUUCAUC---------AUCAUCUAGGGUUUCCUCUCCUGAUCCGACCAAAUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUG ....((((.....(((---------(......(((....)))....))))..))))......((((((((((.(.......).))))))))))............... ( -26.80) >DroPse_CAF1 11015 108 - 1 GAGCGGUUAGCUCUUCGUCAUCGUUGUCAUCCAAUGUCUCCUCACCUGAUCCGACUAGUUUUGCCCCCGGGUGUCCUACUGUGACCCGGGGGUUUUCAAUAAAAAAUG ((((.....))))...(((..(((((.....))))).....((....))...)))..(....((((((((((...........))))))))))...)........... ( -30.80) >DroSec_CAF1 14679 99 - 1 CAGCGGUUAUUUCAUC---------AUCAUCAAGGGUUUCCUCUCCUGAUCCGACCAAAUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUG ....((((........---------...((((.(((......))).))))..))))......((((((((((.(.......).))))))))))............... ( -27.22) >DroYak_CAF1 7044 105 - 1 CAGCGGUUAUUUCAUC---AUCAUCAUCAUCUAGGGUUUCCUCUCCUGAUCCGACCAACUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUG ....((((...(((..---......(((......))).........)))...))))......((((((((((.(.......).))))))))))............... ( -25.93) >DroAna_CAF1 83192 99 - 1 CACCAGUGAUUUCUUC---------AUCGUCUAUAGUCUCCUCUCCUGAUCCGACUAAUUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUG .....((((.....))---------))......(((((...((....))...))))).....((((((((((.(.......).))))))))))............... ( -27.40) >DroPer_CAF1 16133 108 - 1 GAGCGGUUAGCUCUUCGUCAUCGUUGUCAUCCAAUGUCUCCUCACCAGAUCCGACUAGUUUUGCCCCCGGGUGUCCUACUGUGACCCGGGGGUUUCCAAUAAAAAAUG ((((.....))))...(((..(((((.....)))))(((.......)))...))).......((((((((((...........))))))))))............... ( -30.90) >consensus CAGCGGUUAUUUCAUC_________AUCAUCUAGGGUCUCCUCUCCUGAUCCGACCAAUUUUGCCCCCGGGUGACCUACUUUGACCCGGGGGUUUUCAAUAAAAAAUG ..................................((((...((....))...))))......((((((((((...........))))))))))............... (-23.32 = -23.23 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:15 2006