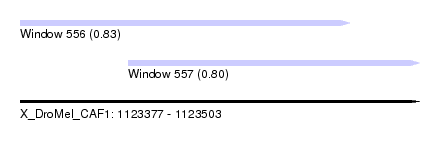
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,123,377 – 1,123,503 |
| Length | 126 |
| Max. P | 0.825778 |

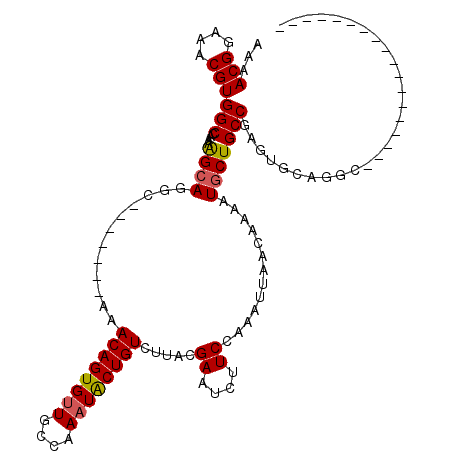
| Location | 1,123,377 – 1,123,481 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1123377 104 + 22224390 ----AAAACAGCGACAGCAAACAA--AUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAAGCAGGC-------AAACAGUGUUGCCAAAUACUGUCUAACGAAUCUUCCUAAUUAA ----......((((..........--...))))..........(((....))).(((.......)))-------..((((((((....))))))))..................... ( -19.92) >DroSec_CAF1 20120 104 + 1 ----AAAACAGCGACAGCAAACAA--AUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAGGCAAGC-------AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAA ----......((....))......--...((((..........(((....))).(((...)))..))-------..((((((((....)))))))).....)).............. ( -20.20) >DroSim_CAF1 21066 104 + 1 ----AAAACAGCGACAGCAAACAA--AUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAAGCAAGC-------AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAA ----......((....))......--...((((..........(((....))).((.....))..))-------..((((((((....)))))))).....)).............. ( -18.00) >DroEre_CAF1 21200 104 + 1 ---AAAAACAGAGCCAGCAAACAA--AUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAAGCAGAC-------AAACAGUGUUGCCAAAUGCUGUCUUACGAAU-UUCCAAAUUAA ---.......(((.(((((.....--.................(((....)))(((........(((-------(.....)))))))...))))).)))......-........... ( -17.60) >DroYak_CAF1 20710 116 + 1 ACAAAAAACAGAGCCAGCAAACAAAAAUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAAGCAGGCAAACGGCAAACAGUGUUGCCAAUUACUGUCUUGCGAAU-UUCCAAAUUAA ..........(.(((.((.............))..........(((....)))))))....(((((((...(((((......)))))......)).)))))....-........... ( -21.92) >consensus ____AAAACAGCGACAGCAAACAA__AUAUCGCACAAAAUAAAACGGAAACGUGGCCAAAAGCAGGC_______AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAA .............................(((...........(((....))).((.....)).............((((((((....))))))))....))).............. (-13.92 = -13.96 + 0.04)

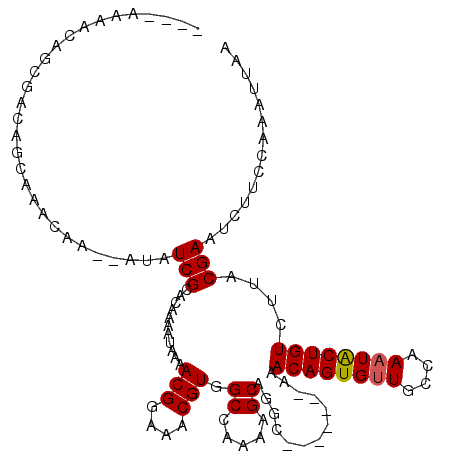
| Location | 1,123,411 – 1,123,503 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1123411 92 + 22224390 AAAACGGAAACGUGGCCAAAAGCAGGC-------AAACAGUGUUGCCAAAUACUGUCUAACGAAUCUUCCUAAUUAACAAAAUUCUGCCGAGUUUAGGC------------------ ...(((....))).(((..((((.(((-------(..(((((((....)))))))((....))......................))))..)))).)))------------------ ( -23.30) >DroSec_CAF1 20154 92 + 1 AAAACGGAAACGUGGCCAAAGGCAAGC-------AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAACAAAAUGCUGCCGAGUGCAGGC------------------ ...(((....))).((....((((.((-------(.((((((((....)))))))).....((....)).............)))))))....))....------------------ ( -23.20) >DroSim_CAF1 21100 92 + 1 AAAACGGAAACGUGGCCAAAAGCAAGC-------AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAACAAAAUGCUGCCGAGUGCAGUC------------------ .....((((.((((((.....))....-------..((((((((....))))))))..))))....)))).............(((((.....))))).------------------ ( -19.30) >DroEre_CAF1 21235 109 + 1 AAAACGGAAACGUGGCCAAAAGCAGAC-------AAACAGUGUUGCCAAAUGCUGUCUUACGAAU-UUCCAAAUUAACAAAAUGCUGCCAAGUGCAGGCGAAUCUACUUCACUUGUC ...(((....)))........((((.(-------(.((((((((....))))))))......(((-(....)))).......))))))((((((.((.........)).)))))).. ( -23.30) >DroYak_CAF1 20750 102 + 1 AAAACGGAAACGUGGCCAAAAGCAGGCAAACGGCAAACAGUGUUGCCAAUUACUGUCUUGCGAAU-UUCCAAAUUAACAAAGUGCUGCCAACUGCAGGCGAAU-------------- ...(((....))).(((...((..((((.((.((((((((((........)))))).)))).(((-(....))))......))..))))..))...)))....-------------- ( -27.40) >consensus AAAACGGAAACGUGGCCAAAAGCAGGC_______AAACAGUGUUGCCAAAUACUGUCUUACGAAUCUUCCAAAUUAACAAAAUGCUGCCGAGUGCAGGC__________________ ...(((....)))(((....((((............((((((((....)))))))).....((....)).............)))))))............................ (-16.74 = -16.82 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:48 2006