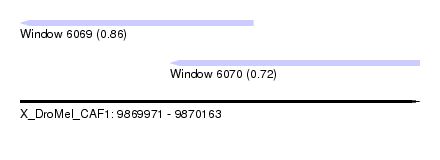
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,869,971 – 9,870,163 |
| Length | 192 |
| Max. P | 0.858125 |

| Location | 9,869,971 – 9,870,083 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9869971 112 - 22224390 UUUCGAACCAUCUUUGGAUCUUAGAUCUCAACUCGCAUCUUCUCGUGAUCAAUGUCAAUGCCCAUACAUUCAUUUACUU-----CACGGCAACAU--CAUCAUCAU-UGCCUGCAUCGAU ..((((.......(((((((...)))).)))...(((.......((((.....((.((((......)))).)).....)-----)))(((((...--........)-))))))).)))). ( -19.90) >DroSec_CAF1 14674 109 - 1 UUUCGGUCCAUCUUUGGAUCUCAGAUCUCAACACGCAUCUUCUCGUGAUCAAUGUCAAUGCUCAUACAUUAAUUUACUA-----CUUGGCAACAU--CAUCA---U-CGCCUGCAUCGAU ....((((((....))))))...........((((........))))(((.((((.((((......)))).........-----...(((.....--.....---.-.))).)))).))) ( -17.10) >DroYak_CAF1 14311 111 - 1 UUUCGAUCCAUCUUUGGAUCACAACU------UCGCAUCUUCUCGUGAUCAAUGUCAAUGCCCAUACAUUCAUUUACUAUGCCACUCGGCAGCAGGCGAUGA---UGUGCCUGCAUCGAU ....((((((....))))))......------((((........)))).....(((.((((...(((((.((((..((.((((....))))..))..)))))---))))...)))).))) ( -30.70) >consensus UUUCGAUCCAUCUUUGGAUCUCAGAUCUCAACUCGCAUCUUCUCGUGAUCAAUGUCAAUGCCCAUACAUUCAUUUACUA_____CUCGGCAACAU__CAUCA___U_UGCCUGCAUCGAU ..((((((((....))))))..............(((.......((((..(((((..........)))))...))))..........(((..................))))))...)). (-15.66 = -15.77 + 0.11)


| Location | 9,870,043 – 9,870,163 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.73 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9870043 120 - 22224390 GUGAAAAAUGGAGACAGUGCCUGUCAAGGCCGGGUACACACACGCCAAUAAAAUAGAAAACCCCACUCGAAAUCAAACGAUUUCGAACCAUCUUUGGAUCUUAGAUCUCAACUCGCAUCU (((.....((....))(((((((.......))))))).)))..((.....................((((((((....)))))))).......(((((((...)))).)))...)).... ( -28.60) >DroSec_CAF1 14743 105 - 1 GUGAAAAAUGGAGACAGUGCCUGUCAAGGC---------------CAAUAAAAUAGAAAACGCCACUCCAAAUCAAACGAUUUCGGUCCAUCUUUGGAUCUCAGAUCUCAACACGCAUCU (((.....(((((((((...)))))..(((---------------................)))..))))........(((((.((((((....))))))..)))))....)))...... ( -25.09) >DroYak_CAF1 14388 110 - 1 GUGAAAAAUAGAGACAGUGCCUGUCAAGGCCGGGUAC----ACGCCAAUAAAAUAGAAAACCCCACUCCAAAUCAAACGAUUUCGAUCCAUCUUUGGAUCACAACU------UCGCAUCU (((((.....(((...(.((((....)))))((((..----..................))))..))).(((((....))))).((((((....)))))).....)------)))).... ( -25.85) >consensus GUGAAAAAUGGAGACAGUGCCUGUCAAGGCCGGGUAC____ACGCCAAUAAAAUAGAAAACCCCACUCCAAAUCAAACGAUUUCGAUCCAUCUUUGGAUCUCAGAUCUCAACUCGCAUCU ((((......(((.....((((....))))..(((........)))...................))).(((((....))))).((((((....))))))............)))).... (-16.79 = -17.73 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:08 2006