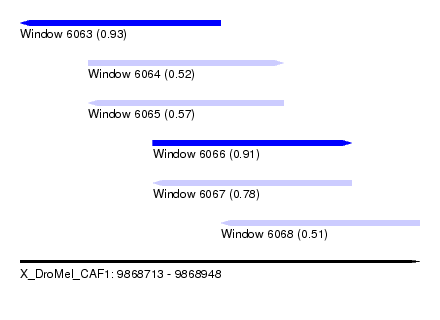
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,868,713 – 9,868,948 |
| Length | 235 |
| Max. P | 0.933815 |

| Location | 9,868,713 – 9,868,831 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -20.50 |
| Energy contribution | -22.50 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868713 118 - 22224390 GUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCUACGAA-AAUGUCAUUA-AAAAAACGGCAACAUUUAAAGUUUUAAUUGAAAGAUAAGAUAGCAAAGUGUUGCUGAUUUUAU .((((((....))))))(.((.....)).)(((((.((.......-.)).))))).-.((((.(((((((((((...((((((.((....))))))))....))))))))))).)))).. ( -29.30) >DroSec_CAF1 13435 119 - 1 GUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA-AAAAAACGGCAACUUUUAAGCUUUUAAUUAAAAGAUAAGAUAGCAACGUGUUGCUGAUUUUAU .((((((....))))))((((.....))..((((((((.........)))))))).-........))..(((((((........)))))))((((((((((((....))))).))))))) ( -27.10) >DroSim_CAF1 13598 119 - 1 GUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA-AAAAAACGGCAACUUUUAAGCUUUUAAUUAAAAGAUAAGAUAGCAAAGUGUUGCUGAUUUUAU .((((((....))))))((((.....))..((((((((.........)))))))).-........))..(((((((........)))))))((((((((((((....))))).))))))) ( -25.90) >DroYak_CAF1 13033 107 - 1 GUUUGGCGAAAGUCAAAGCGGCUAAGCCACGA-------------AAAUUGCAUUACAAAAAACGGCAGCUCUUAACUUUUUAAUUGAAAGAUAAGAUAGCAAAGUGUUGCUGAUUUUAU .((((((....))))))(.(((...))).)..-------------.............((((.(((((((.(((..((.(((.(((....))).))).))..))).))))))).)))).. ( -27.30) >consensus GUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA_AAAAAACGGCAACUUUUAAGCUUUUAAUUAAAAGAUAAGAUAGCAAAGUGUUGCUGAUUUUAU .((((((....))))))((((.....))..((((((((.........))))))))..........))..(((((((........)))))))((((((((((((....))))).))))))) (-20.50 = -22.50 + 2.00)


| Location | 9,868,753 – 9,868,868 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868753 115 + 22224390 AACUUUAAAUGUUGCCGUUUUUU-UAAUGACAUU-UUCGUAGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---CUCCACGCUUGCC ......((((((...((((....-.)))))))))-)...((((((.......))))))((((((((.......((((......))))..)))))))).......---............. ( -23.30) >DroSec_CAF1 13475 116 + 1 AAGCUUAAAAGUUGCCGUUUUUU-UAAUGAAAUUUUUUGUGGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---UUCCACGCUUGCC ((((..(((((((..((((....-.)))).))))))).(((((((.......))....((((((((.......((((......))))..)))))))).......---.)))))))))... ( -30.00) >DroSim_CAF1 13638 116 + 1 AAGCUUAAAAGUUGCCGUUUUUU-UAAUGAAAUUUUUUGUGGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---CACCACGCUUGCC ((((..(((((((..((((....-.)))).))))))).(((((((.......)))...((((((((.......((((......))))..)))))))).......---..))))))))... ( -30.10) >DroYak_CAF1 13073 107 + 1 AAAGUUAAGAGCUGCCGUUUUUUGUAAUGCAAUUU-------------UCGUGGCUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCCCCACUCCACACUUGCC .((((...((((..(.(....(((.....)))...-------------.))..)))).((((((((.......((((......))))..))))))))................))))... ( -25.10) >consensus AAGCUUAAAAGUUGCCGUUUUUU_UAAUGAAAUUUUUUGUGGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC___CUCCACGCUUGCC ............((((..................................(((((...)))))....(((((.((((......))))..)))))))))...................... (-19.31 = -19.12 + -0.19)

| Location | 9,868,753 – 9,868,868 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -24.84 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868753 115 - 22224390 GGCAAGCGUGGAG---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCUACGAA-AAUGUCAUUA-AAAAAACGGCAACAUUUAAAGUU ((((..(((((((---(.((....))...(((((((.(...((((((....)))))).)))))))))).(.....).))))))..-..))))....-.......(....).......... ( -29.70) >DroSec_CAF1 13475 116 - 1 GGCAAGCGUGGAA---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA-AAAAAACGGCAACUUUUAAGCUU ...((((...(((---((....((((...(((((((.(...((((((....)))))).))))))))....((((((((.........)))))))).-.......)))).)))))..)))) ( -32.90) >DroSim_CAF1 13638 116 - 1 GGCAAGCGUGGUG---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA-AAAAAACGGCAACUUUUAAGCUU ...((((....((---((((..((((...(((((((.(...((((((....)))))).))))))))....((((((((.........)))))))).-.......)))).)))))).)))) ( -32.70) >DroYak_CAF1 13073 107 - 1 GGCAAGUGUGGAGUGGGGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAGCCACGA-------------AAAUUGCAUUACAAAAAACGGCAGCUCUUAACUUU .((((.((((((((((.(....).)))))(((((((.(...((((((....)))))).))))))))))))).-------------...))))............................ ( -32.60) >consensus GGCAAGCGUGGAG___GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCCACAAAAAAUUUCAUUA_AAAAAACGGCAACUUUUAAGCUU .((....((((.....((......))...(((((((.(...((((((....)))))).))))))))))))((((((((.........))))))))..........))............. (-24.84 = -26.40 + 1.56)


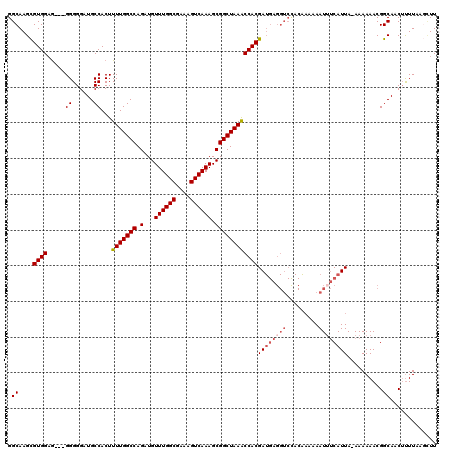
| Location | 9,868,791 – 9,868,908 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868791 117 + 22224390 AGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---CUCCACGCUUGCCAAUCUCUAUCGCCGGGCAAUUUGCGUUAAUGGGGAAAAUU (((((.......))))).((((((((.......((((......))))..)))))))).(((((.---....(((((((((...(......)...)))))...))))....)))))..... ( -37.90) >DroSec_CAF1 13514 117 + 1 GGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---UUCCACGCUUGCCAAUCUCUAUCGCCGGGCAAUUUGCGUUAAUGGGGAAAAUU (((((.......))))).((((((((.......((((......))))..)))))))).(((((.---....(((((((((...(......)...)))))...))))....)))))..... ( -37.80) >DroSim_CAF1 13677 117 + 1 GGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC---CACCACGCUUGCCAAUCUCUAUCGCCGGGCAAUUUGCGUUAAUGGGGAAAAUU (((((.......))))).((((((((.......((((......))))..))))))))....(((---((..(((((((((...(......)...)))))...))))...)))))...... ( -38.20) >DroYak_CAF1 13108 112 + 1 --------UCGUGGCUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCCCCACUCCACACUUGCCAAUCUCUAUCGCCGGGCAAUUUGCGUUAAUGGGGAAAAUU --------..........((((((((.......((((......))))..)))))))).....(((((...(.((.(((((...(......)...)))))..)).)....)))))...... ( -29.40) >consensus GGACCUCAUCGUGGUUUAGCCGCUUUGACUUUCGCCAAACAUCUGGCCAAAAGUGGCAUCCCCC___CUCCACGCUUGCCAAUCUCUAUCGCCGGGCAAUUUGCGUUAAUGGGGAAAAUU ..................((((((((.......((((......))))..)))))))).(((((........(((((((((...(......)...)))))...))))....)))))..... (-30.70 = -30.95 + 0.25)



| Location | 9,868,791 – 9,868,908 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -34.90 |
| Energy contribution | -34.52 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868791 117 - 22224390 AAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAG---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCU ....((((((....((((...(((((...(......)...)))))))))....---)))))).(((...(((((((.(...((((((....)))))).)))))))).(.....).))).. ( -38.20) >DroSec_CAF1 13514 117 - 1 AAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAA---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCC ....((((((.((.((((...(((((...(......)...))))))))).)).---)))))).(((...(((((((.(...((((((....)))))).)))))))).(.....).))).. ( -37.50) >DroSim_CAF1 13677 117 - 1 AAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGUG---GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCC ...(..((((((((.(((...(((((...(......)...)))))))))))))---)))..).(((...(((((((.(...((((((....)))))).)))))))).(.....).))).. ( -40.90) >DroYak_CAF1 13108 112 - 1 AAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGUGUGGAGUGGGGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAGCCACGA-------- .(((..(((((((.((((...(((((...(......)...)))))))))..)))))))..)))((((....))))......((((((....))))))(.(((...))).)..-------- ( -42.00) >consensus AAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAG___GGGGGAUGCCACUUUUGGCCAGAUGUUUGGCGAAAGUCAAAGCGGCUAAACCACGAUGAGGUCC ....((((((....((((...(((((...(......)...))))))))).......)))))).......(((((((.(...((((((....)))))).)))))))).............. (-34.90 = -34.52 + -0.38)



| Location | 9,868,831 – 9,868,948 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -33.87 |
| Energy contribution | -34.36 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9868831 117 - 22224390 GGCCGCUUAUCACACUGCAUUUGUCGCCGUUUAAUGGUGCAAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAG---GGGGGAUGCCACUUUUGGCCAGAU ((((......(((.((((.((((((((((.(((((((.(........)))))))).((.....)).)))))))))).....)).)).)))(((---((((....)).))))))))).... ( -39.50) >DroSec_CAF1 13554 117 - 1 AGCCGCUUAUCACUUUGCAUUUGUCGCCGUUUAAUGGUGCAAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAA---GGGGGAUGCCACUUUUGGCCAGAU ..((.(((.(((((((((.((((((((((.(((((((.(........)))))))).((.....)).)))))))))).....))))).)))).)---)).))..((((....))))..... ( -39.70) >DroSim_CAF1 13717 117 - 1 AGCCGCUUAUCACUUUGCAUUUGUCGCCGUUUAAUGGUGCAAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGUG---GGGGGAUGCCACUUUUGGCCAGAU ..((.(((((((((((((.((((((((((.(((((((.(........)))))))).((.....)).)))))))))).....))))).))))))---)).))..((((....))))..... ( -43.70) >DroYak_CAF1 13140 120 - 1 GGCCGCUUAUCACUUUGCAUUUGUCGCCGUUUAAUGCUGCAAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGUGUGGAGUGGGGGGGAUGCCACUUUUGGCCAGAU ((((......((((((((.((((((((((..((((..(((................))).))))..)))))))))).....))))).)))((((((.(....).))))))..)))).... ( -43.09) >consensus AGCCGCUUAUCACUUUGCAUUUGUCGCCGUUUAAUGGUGCAAUUUUCCCCAUUAACGCAAAUUGCCCGGCGAUAGAGAUUGGCAAGCGUGGAG___GGGGGAUGCCACUUUUGGCCAGAU ..((.((..(((((((((.((((((((((..((((..(((................))).))))..)))))))))).....))))).)))).....)).))..((((....))))..... (-33.87 = -34.36 + 0.50)


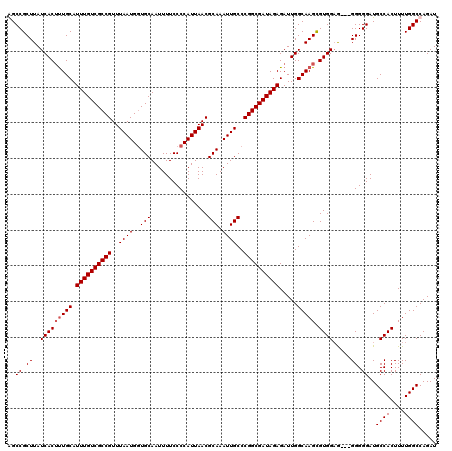
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:07 2006