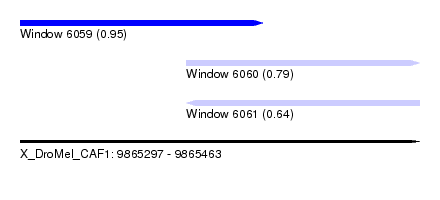
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,865,297 – 9,865,463 |
| Length | 166 |
| Max. P | 0.951274 |

| Location | 9,865,297 – 9,865,398 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -24.13 |
| Energy contribution | -25.38 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9865297 101 + 22224390 UACCGGUUU-UUGGGCUAUUAAACCGCAUUGGCCUGAAAUUCCAGCAUUCCCAG----------UGCAAUCGCCUUUGGUAAUUGUAUCUGUCAGAUGCACAGAUACACGUG-------- ((((((.((-(..(((((...........)))))..)))..)).((((.....)----------)))..........))))..((((((((((....).)))))))))....-------- ( -31.40) >DroPse_CAF1 10208 120 + 1 UACCGGUUUUUUGUGCUAUUAAACCGCAUUUGCCUGAAAUUCCAGCAUUCCCAGUCGCAUCCACUGCAAUCGCUUUUGGUAACUGUAUCUGACAGAUACACAGAUACACCGGGCCACAAA ..(((((..((((((.((((..........(((.((......)))))....(((..(((.(((..((....))...)))....)))..)))...))))))))))...)))))........ ( -26.80) >DroSec_CAF1 10070 102 + 1 UACCGGUUUUUUGGGCUAUUAAACCGCAUUGGCCUGAAAUUCCAGCAUUCCCAG----------UGCAAUCGCCUUUGGUAAUUGUAUCUGUCAGAUGCACAGAUACACGUG-------- ((((((..(((..(((((...........)))))..)))..)).((((.....)----------)))..........))))..((((((((((....).)))))))))....-------- ( -33.40) >DroYak_CAF1 9521 101 + 1 UACCGGUUU-UUGGGCUAUUAAACCGCAUUGGCCUGAAAUUCCAGCAUUCCCAG----------UGCAAUCGCUUUUGGUAAUUGUAUCUGUCAGAUGCACAGAUACACGUG-------- ((((((.((-(..(((((...........)))))..)))..)).((((.....)----------)))..........))))..((((((((((....).)))))))))....-------- ( -31.40) >consensus UACCGGUUU_UUGGGCUAUUAAACCGCAUUGGCCUGAAAUUCCAGCAUUCCCAG__________UGCAAUCGCCUUUGGUAAUUGUAUCUGUCAGAUGCACAGAUACACGUG________ ((((((..((((((((((...........))))))))))..)).(((.................)))..........))))..(((((((((.......)))))))))............ (-24.13 = -25.38 + 1.25)


| Location | 9,865,366 – 9,865,463 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.86 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9865366 97 + 22224390 AAUUGUAUCUGUCAGAUGCACAGAUACACGUG----------CA-UCGGGAUCUGUCUUCAGUUUUAUUGCAAAAUGUGCCAUUUUUCUUUUUUAUUUUUGUUGGCUA ...((((((((((....).)))))))))..((----------((-..((((.(((....)))))))..))))......((((....................)))).. ( -21.55) >DroPse_CAF1 10288 108 + 1 AACUGUAUCUGACAGAUACACAGAUACACCGGGCCACAAAUACACACGUGCGGUGCCUUCAACCUCAUUGCAAAAUUUGCUACAUUUUUUUUUGUAUUUUGUCUUCCU ...((((((((.........))))))))..(((..(((((((((...(((.(((.......))).))).(((.....)))............)))).)))))..))). ( -22.30) >DroSec_CAF1 10140 77 + 1 AAUUGUAUCUGUCAGAUGCACAGAUACACGUG----------CA-UCGGGAUCUGUCUUCAGUUUUAUUGCAAAAUGUGCCAUUUUUU-------------------- ...((((((((((....).)))))))))..((----------((-..((((.(((....)))))))..))))................-------------------- ( -17.70) >consensus AAUUGUAUCUGUCAGAUGCACAGAUACACGUG__________CA_UCGGGAUCUGUCUUCAGUUUUAUUGCAAAAUGUGCCAUUUUUUUUUUU___UUUUGU___C__ ...(((((((((.......))))))))).........................................(((.....)))............................ ( -9.90 = -10.23 + 0.33)


| Location | 9,865,366 – 9,865,463 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 64.86 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9865366 97 - 22224390 UAGCCAACAAAAAUAAAAAAGAAAAAUGGCACAUUUUGCAAUAAAACUGAAGACAGAUCCCGA-UG----------CACGUGUAUCUGUGCAUCUGACAGAUACAAUU ..((((....................))))......((((......(((....))).......-))----------))..(((((((((.......)))))))))... ( -18.07) >DroPse_CAF1 10288 108 - 1 AGGAAGACAAAAUACAAAAAAAAAUGUAGCAAAUUUUGCAAUGAGGUUGAAGGCACCGCACGUGUGUAUUUGUGGCCCGGUGUAUCUGUGUAUCUGUCAGAUACAGUU .(((.(((....((((........))))(((.....)))......)))....((((((((((........))))...)))))).))).((((((.....))))))... ( -21.90) >DroSec_CAF1 10140 77 - 1 --------------------AAAAAAUGGCACAUUUUGCAAUAAAACUGAAGACAGAUCCCGA-UG----------CACGUGUAUCUGUGCAUCUGACAGAUACAAUU --------------------..((((((...))))))(((......(((....))).......-))----------)...(((((((((.......)))))))))... ( -14.42) >consensus __G___ACAAAA___AAAAAAAAAAAUGGCACAUUUUGCAAUAAAACUGAAGACAGAUCCCGA_UG__________CACGUGUAUCUGUGCAUCUGACAGAUACAAUU ............................(((.....))).........................................(((((((((.......)))))))))... (-11.40 = -11.73 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:00 2006