
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,857,726 – 9,857,846 |
| Length | 120 |
| Max. P | 0.819418 |

| Location | 9,857,726 – 9,857,846 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.72 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506752 |
| Prediction | RNA |
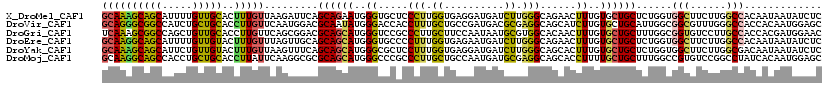
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9857726 120 + 22224390 GAGAUAUUAUUGUGGCCAAGAAGCCACCAGAGCAGCACAAAGUUCUGCCCAAGAUCAUCCUCACCAAGGGAGCACCCAUUCUGCUGAAUCUUAACAAAGUGCAACAAAAUGCUGCUUUGC ...........(((((......)))))((((((((((....(.(((.....))).).((((......))))((((..((((....)))).........)))).......)))))))))). ( -31.10) >DroVir_CAF1 2971 120 + 1 GCUCCAUUGUGGUGGCCCAAACGCCGCCAAUGCAGCACAAGAUGCUGCCUCGCGUCAUCGGCAGCAAAGGUGGUCCCAUAUUGCGUCCAUUGAACAAGGUGCAGCAGAUGGCCGCCCUGC (((.(((..(((((((......)))))))))).)))......(((((((..........)))))))..(((((..((((.((((...((((......))))..))))))))))))).... ( -45.90) >DroGri_CAF1 8327 120 + 1 GUUCCAUCGUGGUGGCAAGGACACCGCCAAAGCAGCACAAAGUUGUGCCACGCAUUAUUGGAAGCAAGGGCGGACCCAUGCUGCGUCCGCUGAACAAGGUGCAACAGCUGGCCGCUUUGA ........((((((.......))))))((((((.((.((..((((..((........(((....))).(((((((.(.....).)))))))......))..))))...)))).)))))). ( -45.20) >DroEre_CAF1 2498 120 + 1 GAGAUAUUAUUGUGGCCAAGAAGCCACCAGAGCAGCACAAAGUUCUGCCCAAGAUCAUUCUCACCAAAGGGGCACCCAUGCUGCUGCAACUAAACAAAGUACAACAAAAUGCUGCCUUGC ((((.......(((((......)))))((((((........))))))...........))))....((((((((....(((....)))(((......))).........)))).)))).. ( -28.20) >DroYak_CAF1 1930 120 + 1 GAGAUAUUAUUGUCGCCAAGAAGCCACCAGAGCAGCACAAAGUGCUGCCCAAGAUCAUCCUCACCAAAGGAGCGCCCAUGCUGCUGAAACUUAACAAAGUACAACAGAAUGCUGCUUUGC (((........((.((......)).))..(.(((((((...))))))).).........)))..(((((.((((......(((.((..((((....)))).)).)))..)))).))))). ( -27.90) >DroMoj_CAF1 2857 120 + 1 GCUCCAUUGUGAUAGGCCGGACACGGCCAAAGCAGCAAAAGGUGCUGCCUCGCAUCAUUGGCAGCAAGGGCGGGCCCAUGCUGCGCGCCUUGAAUAAGGUGCAGCAGGUGGCUGCCUUGC ((((((.(((((..(((((....)))))...((((((.....)))))).)))))....))).))).(((((((.(((.((((((..(((((....))))))))))))).).))))))).. ( -61.60) >consensus GAGACAUUAUGGUGGCCAAGAAGCCACCAAAGCAGCACAAAGUGCUGCCCAACAUCAUCCGCACCAAAGGAGCACCCAUGCUGCGGCAACUGAACAAAGUGCAACAGAAGGCUGCCUUGC ...........(((((......)))))....((((((...............((....))........((.....)).))))))..........((((((((........).))))))). (-15.54 = -15.40 + -0.14)

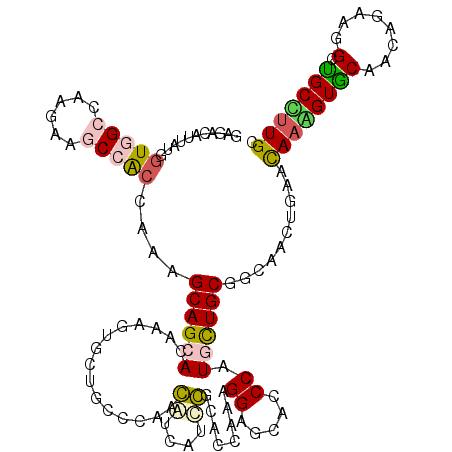

| Location | 9,857,726 – 9,857,846 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.72 |
| Mean single sequence MFE | -44.37 |
| Consensus MFE | -21.50 |
| Energy contribution | -20.25 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9857726 120 - 22224390 GCAAAGCAGCAUUUUGUUGCACUUUGUUAAGAUUCAGCAGAAUGGGUGCUCCCUUGGUGAGGAUGAUCUUGGGCAGAACUUUGUGCUGCUCUGGUGGCUUCUUGGCCACAAUAAUAUCUC .((.((((((((....((((.((..(...(..((((.(((...(((....)))))).))))..)...)..))))))......)))))))).))((((((....))))))........... ( -39.20) >DroVir_CAF1 2971 120 - 1 GCAGGGCGGCCAUCUGCUGCACCUUGUUCAAUGGACGCAAUAUGGGACCACCUUUGCUGCCGAUGACGCGAGGCAGCAUCUUGUGCUGCAUUGGCGGCGUUUGGGCCACCACAAUGGAGC .((.((.((((...((((((..((((((((.(((..((((...((.....)).))))..))).))).)))))(((((((...)))))))....))))))....)))).))....)).... ( -49.20) >DroGri_CAF1 8327 120 - 1 UCAAAGCGGCCAGCUGUUGCACCUUGUUCAGCGGACGCAGCAUGGGUCCGCCCUUGCUUCCAAUAAUGCGUGGCACAACUUUGUGCUGCUUUGGCGGUGUCCUUGCCACCACGAUGGAAC .....(((.((.((((..((.....)).)))))).)))((((.(((....))).))))((((.......(..(((((....)))))..)..(((.(((......))).)))...)))).. ( -44.90) >DroEre_CAF1 2498 120 - 1 GCAAGGCAGCAUUUUGUUGUACUUUGUUUAGUUGCAGCAGCAUGGGUGCCCCUUUGGUGAGAAUGAUCUUGGGCAGAACUUUGUGCUGCUCUGGUGGCUUCUUGGCCACAAUAAUAUCUC .((..(((((....(((((((((......)).)))))))(((.((.(((((.......((((....)))))))))...)).))))))))..))((((((....))))))........... ( -40.41) >DroYak_CAF1 1930 120 - 1 GCAAAGCAGCAUUCUGUUGUACUUUGUUAAGUUUCAGCAGCAUGGGCGCUCCUUUGGUGAGGAUGAUCUUGGGCAGCACUUUGUGCUGCUCUGGUGGCUUCUUGGCGACAAUAAUAUCUC .(((((.(((.(((((((((((((....))))....)))))).))).))).)))))....(((((.....(((((((((...)))))))))..((.(((....))).)).....))))). ( -39.20) >DroMoj_CAF1 2857 120 - 1 GCAAGGCAGCCACCUGCUGCACCUUAUUCAAGGCGCGCAGCAUGGGCCCGCCCUUGCUGCCAAUGAUGCGAGGCAGCACCUUUUGCUGCUUUGGCCGUGUCCGGCCUAUCACAAUGGAGC (((((((.(((.(.((((((.((((....))))...)))))).))))..).))))))..(((.(((((.(((((((((.....)))))))))(((((....))))))))))...)))... ( -53.30) >consensus GCAAAGCAGCAAUCUGUUGCACCUUGUUCAGUGGCAGCAGCAUGGGUCCGCCCUUGCUGACAAUGAUCCGGGGCAGAACUUUGUGCUGCUCUGGCGGCGUCUUGGCCACAACAAUAGAUC ((((((((((.....)))))..))))).........((((((..((.....(((.((..........)).)))......))..))))))......(((......)))............. (-21.50 = -20.25 + -1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:53 2006