
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,844,624 – 9,844,757 |
| Length | 133 |
| Max. P | 0.917050 |

| Location | 9,844,624 – 9,844,726 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -19.51 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9844624 102 - 22224390 GGAUAUAUGUUUUAACUUUAUUCAUU-UUCUAGGGGCCUAAAACCCUGGGCUAAAAGUGUCUGAGUGGAUUGUUAUAUAUAUAUAUAGCAUUACGUAUAUGGA ..((((((((....((((....((((-((((((((........)))))))....)))))...))))....((((((((....))))))))..))))))))... ( -25.40) >DroSec_CAF1 6128 98 - 1 GGAUAUAUGUUUUAACUUUAUUCAUUUUUCUAGGGUCCUA-AACCCUGGGCUAAAAGUGUCUGAGUGGAUUAUUAUAG----AUAUAGCAUUACGUAUAUGGA ..((((((((...................(((((((....-.)))))))(((....(((((((((((...)))).)))----)))))))...))))))))... ( -21.80) >DroSim_CAF1 6386 98 - 1 GGAUAUAUGUUUCAACUUUAUUCAUUUUUCUAGGGGCCUA-AACCCUGGGCUAAAAGUGUCUGAGUGGAUUGUUAUAG----AUAUAGCAUUACGUAUAUGGA ..((((((((.((.((((....(((((((((((((.....-..)))))))...))))))...)))).)).((((((..----..))))))..))))))))... ( -25.10) >DroEre_CAF1 6285 93 - 1 GCAUUUAUGUUU---CUUUAUGCAUU-UUCUAGGGGCCUAAAACCCUGGGCUAAAAGUGUCUGAGUGGAUUGCUAUAA----AUAUUGCAUUACGC--AUGGA ...(((((((..---..(((.(((((-((((((((........)))))))....)))))).)))((((..(((.....----.....)))))))))--))))) ( -20.50) >consensus GGAUAUAUGUUUUAACUUUAUUCAUU_UUCUAGGGGCCUA_AACCCUGGGCUAAAAGUGUCUGAGUGGAUUGUUAUAG____AUAUAGCAUUACGUAUAUGGA ..((((((((......((((((((.....((((((........))))))((.....))...)))))))).(((((((......)))))))..))))))))... (-19.51 = -20.82 + 1.31)



| Location | 9,844,659 – 9,844,757 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.64 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9844659 98 + 22224390 CCACUCAGACACUUUUAGCCCAGGGUUUUAGGCCCCUAGAA-AAUGAAUAAAGUUAAAACAUAUAUCCCUGUUUAUAUGAAAGUGUAUUGAAUUUGAAU ....(((((((((((((...(((((((((((....))))))-..........((....))......)))))......))))))))).))))........ ( -21.90) >DroSec_CAF1 6159 98 + 1 CCACUCAGACACUUUUAGCCCAGGGUU-UAGGACCCUAGAAAAAUGAAUAAAGUUAAAACAUAUAUCCCUGUUUAUAUGAAAGUGUAUUGAAUUUGAAU ....(((((((((((((...(((((((-(((....)))))............((....))......)))))......))))))))).))))........ ( -21.00) >DroSim_CAF1 6417 98 + 1 CCACUCAGACACUUUUAGCCCAGGGUU-UAGGCCCCUAGAAAAAUGAAUAAAGUUGAAACAUAUAUCCCUGUUUAUAUGAAAGUGUAUUGAAUUUGAAU ....(((((((((((((...(((((((-(((....)))))............((....))......)))))......))))))))).))))........ ( -21.00) >DroEre_CAF1 6314 95 + 1 CCACUCAGACACUUUUAGCCCAGGGUUUUAGGCCCCUAGAA-AAUGCAUAAAG---AAACAUAAAUGCCUGUUUAUAUGAAAGUGUAUUGAAUUUGAAU ....(((((((((((((...((((.((((((....))))))-.(((.......---...))).....))))......))))))))).))))........ ( -18.90) >consensus CCACUCAGACACUUUUAGCCCAGGGUU_UAGGCCCCUAGAA_AAUGAAUAAAGUUAAAACAUAUAUCCCUGUUUAUAUGAAAGUGUAUUGAAUUUGAAU ....(((((((((((((...(((((((((((....))))))..(((.............)))....)))))......))))))))).))))........ (-17.90 = -18.64 + 0.75)

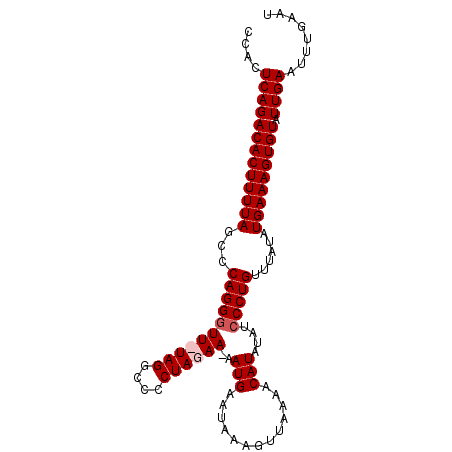
| Location | 9,844,659 – 9,844,757 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9844659 98 - 22224390 AUUCAAAUUCAAUACACUUUCAUAUAAACAGGGAUAUAUGUUUUAACUUUAUUCAUU-UUCUAGGGGCCUAAAACCCUGGGCUAAAAGUGUCUGAGUGG ......(((((..(((((((((((((........)))))).................-.(((((((........)))))))...))))))).))))).. ( -21.60) >DroSec_CAF1 6159 98 - 1 AUUCAAAUUCAAUACACUUUCAUAUAAACAGGGAUAUAUGUUUUAACUUUAUUCAUUUUUCUAGGGUCCUA-AACCCUGGGCUAAAAGUGUCUGAGUGG ......(((((..(((((((((((((........))))))...................((((((((....-.))))))))...))))))).))))).. ( -21.60) >DroSim_CAF1 6417 98 - 1 AUUCAAAUUCAAUACACUUUCAUAUAAACAGGGAUAUAUGUUUCAACUUUAUUCAUUUUUCUAGGGGCCUA-AACCCUGGGCUAAAAGUGUCUGAGUGG ......(((((..(((((((((((((........))))))...................(((((((.....-..)))))))...))))))).))))).. ( -21.30) >DroEre_CAF1 6314 95 - 1 AUUCAAAUUCAAUACACUUUCAUAUAAACAGGCAUUUAUGUUU---CUUUAUGCAUU-UUCUAGGGGCCUAAAACCCUGGGCUAAAAGUGUCUGAGUGG ......(((((..(((((((..((((((.(((((....)))))---.))))))....-.(((((((........)))))))...))))))).))))).. ( -23.50) >consensus AUUCAAAUUCAAUACACUUUCAUAUAAACAGGGAUAUAUGUUUUAACUUUAUUCAUU_UUCUAGGGGCCUA_AACCCUGGGCUAAAAGUGUCUGAGUGG ......(((((..(((((((...........(((((...(......)..))))).....(((((((........)))))))...))))))).))))).. (-21.10 = -20.98 + -0.13)

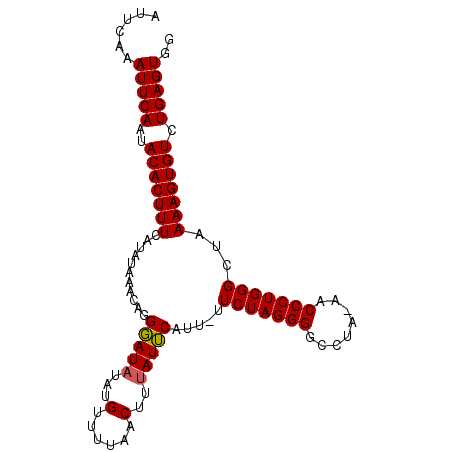

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:50 2006