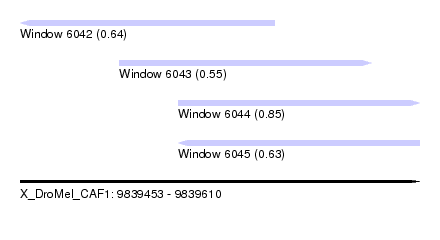
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,839,453 – 9,839,610 |
| Length | 157 |
| Max. P | 0.854560 |

| Location | 9,839,453 – 9,839,553 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -11.77 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9839453 100 - 22224390 AAACAAUGCUAAAAACUGGCCAGCUCUCUGGACGAAUA-----G-UUUCGGCUAACAAAUUAGCA-GGGGGCUGGGUGUACGGGGUGCUAAUUG-CCGACACAGUGAU .......(((.........((((((((((...((((..-----.-.))))(((((....))))))-)))))))))((((.(((.(.......).-))))))))))... ( -34.10) >DroPse_CAF1 8023 95 - 1 AAACAAUGCUAAAA-----------CGCAGACCGAACAAAAGAGUUUUUGGGUAACAAAUUAGCACAGGGGCUGGC--CAGGGAGUGUUAAUUGGCCAGCACAAUGGU ......((((((..-----------.....(((((((......))))...)))......))))))((..(((((((--(((..........))))))))).)..)).. ( -27.12) >DroSec_CAF1 1159 100 - 1 AAACAAUGCUAAAAACUGGCAAGCUCUCUAGCCGAAUA-----G-UUUCGGCUAACAAAUUAGCA-GGGGGCUGGGUGUACGGGGUGCUAAUUG-CCGACACAUUGAU ...((((((((.....)))).(((((((((((((((..-----.-.)))))))...........)-)))))))..((((.(((.(.......).-))))))))))).. ( -32.30) >DroSim_CAF1 1358 100 - 1 AAACAAUGCUAAAAACUGGCCAGCUCUCUGGCCGAAUA-----G-UUUCGGCUAACAAAUUAGCA-GGGGGCUGGGUGUACGGGGUGCUAAUUG-CCGACACAGUGAU .......(((.........(((((((((((((((((..-----.-.))))))(((....))).))-)))))))))((((.(((.(.......).-))))))))))... ( -36.50) >DroEre_CAF1 1147 96 - 1 AAACAAUGCUA--AACUGGCCAGCUCUCUGGCCGAAUA-----G-U-UCGGCUAACAAAUUAGCA-GGGGGCUGGGUGUACGG-GUGCUAAUUG-CCGACACAGUGAU .......(((.--......((((((((((((((((...-----.-.-)))))(((....))).))-)))))))))((((.(((-..........-))))))))))... ( -33.50) >DroPer_CAF1 8178 95 - 1 AAACAAUGCUAAAA-----------CGCAGACCGAACAAAAGAGUUUUUGGGUAACAAAUUAGCACAGGGGCUGGC--CAGGUAGUGUUAAUUGGCCAGCACAAUGGU ......((((((..-----------.....(((((((......))))...)))......))))))((..(((((((--(((..........))))))))).)..)).. ( -27.12) >consensus AAACAAUGCUAAAAACUGGCCAGCUCUCUGGCCGAAUA_____G_UUUCGGCUAACAAAUUAGCA_GGGGGCUGGGUGUACGGGGUGCUAAUUG_CCGACACAGUGAU ...((.((....................((((((((..........))))))))...((((((((.....((.....))......))))))))........)).)).. (-11.77 = -12.30 + 0.53)


| Location | 9,839,492 – 9,839,591 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.25 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9839492 99 + 22224390 CC-UGCUAAUUUGUUAGCCGAAA-C-----UAUUCGUCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUU ..-....((((((((.((((((.-.-----..))))..(((((.(..(((((.......((((......)))).....--..)))))..).))))).)))))))))). ( -22.34) >DroPse_CAF1 8061 97 + 1 CUGUGCUAAUUUGUUACCCAAAAACUCUUUUGUUCGGUCUGCG-----------UUUUAGCAUUGUUUUAUGUCAAUUUCUGUGGCUUUCACUUUGUGCAACAAAUUU ..(((((((..(((.((((((((....)))))...)))..)))-----------..))))))).......((((((.....(((.....))).))).)))........ ( -16.00) >DroSec_CAF1 1198 99 + 1 CC-UGCUAAUUUGUUAGCCGAAA-C-----UAUUCGGCUAGAGAGCUUGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUU ..-....(((((((((((((((.-.-----..))))))).(((((((....))))))).((((.((.....(((((..--.)))))....))...)))))))))))). ( -24.40) >DroSim_CAF1 1397 99 + 1 CC-UGCUAAUUUGUUAGCCGAAA-C-----UAUUCGGCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUU ..-....((((((((.((((((.-.-----..))))))(((((.(..(((((.......((((......)))).....--..)))))..).)))))...)))))))). ( -27.14) >DroEre_CAF1 1185 96 + 1 CC-UGCUAAUUUGUUAGCCGA-A-C-----UAUUCGGCCAGAGAGCUGGCCAGUU--UAGCAUUGUUUUAUGUCAUAU--UUUGGCUUUCACUUUGUGCAACAAAUUU ..-((((((..((..((((((-(-.-----(((..((((((....))))))....--..((((......)))).))).--)))))))..))..))).)))........ ( -27.80) >DroPer_CAF1 8216 97 + 1 CUGUGCUAAUUUGUUACCCAAAAACUCUUUUGUUCGGUCUGCG-----------UUUUAGCAUUGUUUUAUGUCAAUUUCUGUGACUUUCACUUUGUGCAACAAAUUU ..(((((((..(((.((((((((....)))))...)))..)))-----------..))))))).......((((((.....(((.....))).))).)))........ ( -16.90) >consensus CC_UGCUAAUUUGUUAGCCGAAA_C_____UAUUCGGCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU__UAUGGCUUUCACUUUGUGCAACAAAUUU .......((((((((.((((((..........)))))).....................((((.((.....((((.......))))....))...)))))))))))). (-13.39 = -14.25 + 0.86)


| Location | 9,839,515 – 9,839,610 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -15.99 |
| Energy contribution | -17.72 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9839515 95 + 22224390 --UAUUCGUCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUCUCCGAAAUCG --..((((....(((((..(((((.......((((......)))).....--..)))))..)..(((((((((.....))))))))))))))))).... ( -24.64) >DroPse_CAF1 8089 86 + 1 UUUGUUCGGUCUGCG-----------UUUUAGCAUUGUUUUAUGUCAAUUUCUGUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUGUCUGGUCU-- ...........(((.-----------.....))).....................((((..((.(((((((((.....)))))))))))...)))).-- ( -16.80) >DroSec_CAF1 1221 95 + 1 --UAUUCGGCUAGAGAGCUUGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUCUCCGAAAUCG --..(((((..((((((...((((.......((((......)))).....--..)))).)))..(((((((((.....))))))))))))))))).... ( -25.14) >DroSim_CAF1 1420 95 + 1 --UAUUCGGCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU--UAUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUCUCCGAAAUCG --..(((((..((((((..(((((.......((((......)))).....--..))))))))..(((((((((.....))))))))))))))))).... ( -26.14) >DroEre_CAF1 1207 93 + 1 --UAUUCGGCCAGAGAGCUGGCCAGUU--UAGCAUUGUUUUAUGUCAUAU--UUUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUCUCCGAAAUCG --..(((((..((((((..((((((..--((((((......))))..)).--.)))))))))..(((((((((.....))))))))))))))))).... ( -26.80) >DroPer_CAF1 8244 86 + 1 UUUGUUCGGUCUGCG-----------UUUUAGCAUUGUUUUAUGUCAAUUUCUGUGACUUUCACUUUGUGCAACAAAUUUGCACAAAUGUCUGGUCU-- ...........(((.-----------.....))).....................((((..((.(((((((((.....)))))))))))...)))).-- ( -17.40) >consensus __UAUUCGGCCAGAGAGCUGGCCAGUUUUUAGCAUUGUUUUAUGUCAUAU__UAUGGCUUUCACUUUGUGCAACAAAUUUGCACAAAUCUCCGAAAUCG ....(((((...(((((((............((((......))))..........)))))))..(((((((((.....)))))))))...))))).... (-15.99 = -17.72 + 1.72)



| Location | 9,839,515 – 9,839,610 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -11.88 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9839515 95 - 22224390 CGAUUUCGGAGAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCAUA--AUAUGACAUAAAACAAUGCUAAAAACUGGCCAGCUCUCUGGACGAAUA-- ....(((((((((((((((((.....)))))))))((....((((..--....(.(((......)))).......))))..))))))))))......-- ( -23.62) >DroPse_CAF1 8089 86 - 1 --AGACCAGACAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCACAGAAAUUGACAUAAAACAAUGCUAAAA-----------CGCAGACCGAACAAA --....(((...(((((((((.....)))))))))(((.....))).....)))...........(((.....-----------.)))........... ( -14.20) >DroSec_CAF1 1221 95 - 1 CGAUUUCGGAGAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCAUA--AUAUGACAUAAAACAAUGCUAAAAACUGGCAAGCUCUCUAGCCGAAUA-- ....(((((...(((((((((.....)))))))))..((.((((((.--..)))...........(((((.....))))).))).))...)))))..-- ( -22.20) >DroSim_CAF1 1420 95 - 1 CGAUUUCGGAGAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCAUA--AUAUGACAUAAAACAAUGCUAAAAACUGGCCAGCUCUCUGGCCGAAUA-- ....(((((...(((((((((.....))))))))).......))...--...........................((((((....)))))))))..-- ( -23.40) >DroEre_CAF1 1207 93 - 1 CGAUUUCGGAGAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCAAA--AUAUGACAUAAAACAAUGCUA--AACUGGCCAGCUCUCUGGCCGAAUA-- ....(((((...(((((((((.....))))))))).......))...--.....................--....((((((....)))))))))..-- ( -23.40) >DroPer_CAF1 8244 86 - 1 --AGACCAGACAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGUCACAGAAAUUGACAUAAAACAAUGCUAAAA-----------CGCAGACCGAACAAA --.......((.(((((((((.....)))))))))))....((((.(....))))).........(((.....-----------.)))........... ( -15.70) >consensus CGAUUUCGGAGAUUUGUGCAAAUUUGUUGCACAAAGUGAAAGCCAUA__AUAUGACAUAAAACAAUGCUAAAAACUGGCCAGCUCUCUGGCCGAAUA__ ......(((((((((((((((.....)))))))))(((.....))).....................................)))))).......... (-11.88 = -13.38 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:46 2006