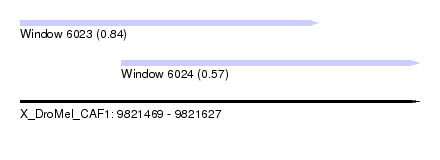
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,821,469 – 9,821,627 |
| Length | 158 |
| Max. P | 0.837996 |

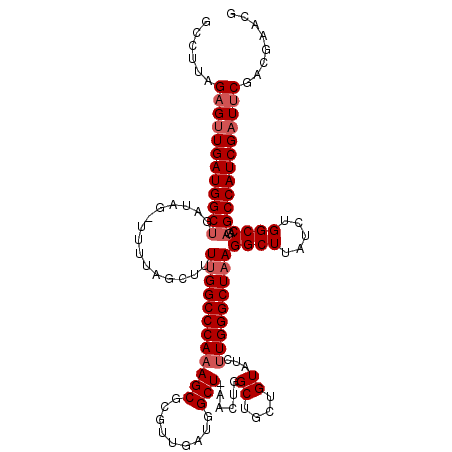
| Location | 9,821,469 – 9,821,587 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -30.48 |
| Energy contribution | -32.40 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
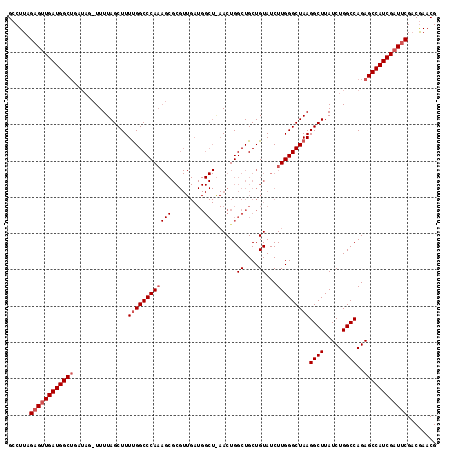
>X_DroMel_CAF1 9821469 118 + 22224390 GCCUUAGAGUUGAUGGCUGAUAGCUUUUAGCUUUUGGCCCAAAGCGCGUUGAUGGCUAAACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACGAACG (((..((((((((.(((.....))).)))))))).))).....(((.((((((((((...((((((...(.....(((((....))))).).)))))))))))))))).)).)..... ( -42.60) >DroSec_CAF1 22902 117 + 1 GCCUUAGAGUUGAUGGCUGAUAGAUUUUAGCUUUUGGCCCAAAGCGCGUUGAUGGCUGAACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUCCG-CGAACG ......(.((..(.((((((......)))))).)..)).)...(((.((((((((((...((((((...(.....(((((....))))).).)))))))))))))))).))-)..... ( -42.50) >DroEre_CAF1 14243 93 + 1 GGCUUAGAGCUGAUGGC-------UUUUAGCUUUCGGCCCA------------------ACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACAAACG ......(((.(((((((-------(((........((((((------------------(...((....))...)))))))..((((.....)))))))))))))).)))........ ( -34.30) >consensus GCCUUAGAGUUGAUGGCUGAUAG_UUUUAGCUUUUGGCCCAAAGCGCGUUGAUGGCU_AACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACGAACG ......((((((((((((...............((((((((((((.........)))......((....))...)))))))))((((.....))))..))))))))))))........ (-30.48 = -32.40 + 1.92)

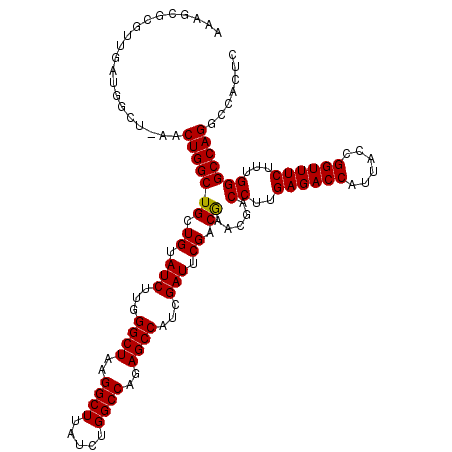
| Location | 9,821,509 – 9,821,627 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -35.22 |
| Energy contribution | -35.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9821509 118 + 22224390 AAAGCGCGUUGAUGGCUAAACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACGAACGACCUUGAGACCGUUACCGGUUUCUUUGGGCCAGGCCACUC ...(((.((((((((((...((((((...(.....(((((....))))).).)))))))))))))))).)).)....(.(((.(((((((....)))))))...))).)......... ( -40.40) >DroSec_CAF1 22942 117 + 1 AAAGCGCGUUGAUGGCUGAACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUCCG-CGAACGACCUUGAGACCCUUACCGGUUUCUUUGGGCCAGGCCACUC ...(((.((((((((((...((((((...(.....(((((....))))).).)))))))))))))))).))-)....(.(((.((((((......))))))...))).)......... ( -41.90) >DroEre_CAF1 14276 100 + 1 A------------------ACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACAAACGACCUUGAGACCAUUACCGGUUUCUUUGGGCCAGGCCACUC .------------------.(((((((.((.(((...((((..((((.....))))..))))...))).)).)).....((..((((((......))))))...)))))))....... ( -35.50) >consensus AAAGCGCGUUGAUGGCU_AACUGGCUGCUGUAUCUUGGGCUAAGGCUUAUCUGGCCAGAGCCAUCGAUUCGACGAACGACCUUGAGACCAUUACCGGUUUCUUUGGGCCAGGCCACUC ....................(((((((.((.(((...((((..((((.....))))..))))...))).)).)).....((..((((((......))))))...)))))))....... (-35.22 = -35.00 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:25 2006