
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,818,517 – 9,818,622 |
| Length | 105 |
| Max. P | 0.862083 |

| Location | 9,818,517 – 9,818,622 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
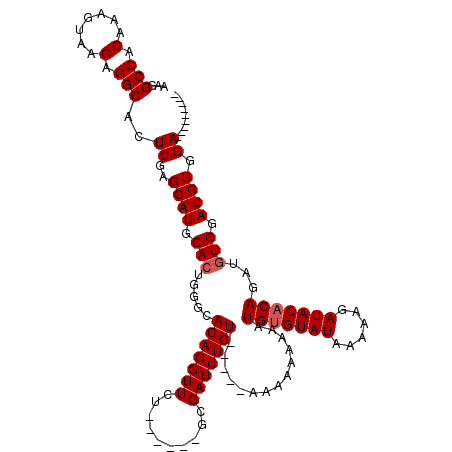
>X_DroMel_CAF1 9818517 105 + 22224390 ---------UCCACAUCCACAUCUGUGUAUCUUUUUAUACACAUUUUUUUUUUUUUAACAGAUCGC------AGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU ---------.(((((..((((((((((.((((.((.....................)).)))))))------)))...))))...((.((((......)))))).......))))).... ( -20.00) >DroSec_CAF1 20000 100 + 1 ---------UCCACAUCCACAUCUGUGUAUCUUUUUAUACACAUUUUUUUUU-----ACAGAUCGC------AGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU ---------.(((((..((((((((((.((((....................-----..)))))))------)))...))))...((.((((......)))))).......))))).... ( -20.05) >DroSim_CAF1 9985 100 + 1 ---------UCCACAUCCACAUCUGUGUAUCUUUUUAUACACAUUUUUUUUU-----ACAGAUCGC------AGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU ---------.(((((..((((((((((.((((....................-----..)))))))------)))...))))...((.((((......)))))).......))))).... ( -20.05) >DroEre_CAF1 11320 97 + 1 ---------UCCACAUCCAUAUCUGUGUAUCUUUUUAUACACAUUUU---UU-----ACAGAUCGC------AGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU ---------.(((((......((((((.((((...............---..-----..)))))))------)))....(((...((.((((......))))))..)))..))))).... ( -18.71) >DroYak_CAF1 11900 112 + 1 GCAGCCACAUCCACAUCCACAUCUGGGUAUCUUUUUAUACACAUUUU---UU-----ACAGAUCGCAGACGCAGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU ...((((((....(((.(((.((((.(.((((...............---..-----..))))).))))(((((...)))))....))).)))...(((((.....)))))))))))... ( -24.51) >consensus _________UCCACAUCCACAUCUGUGUAUCUUUUUAUACACAUUUUUUUUU_____ACAGAUCGC______AGAACCUGUGCCCAGUGCAUGCUCGAGUGCCUCUUACUUUGUGGCCUU .............(((.(((...(((((((......)))))))..............((((.((.........))..)))).....))).)))...(((.(((.(.......).)))))) (-16.72 = -16.76 + 0.04)


| Location | 9,818,517 – 9,818,622 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9818517 105 - 22224390 AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCU------GCGAUCUGUUAAAAAAAAAAAAAUGUGUAUAAAAAGAUACACAGAUGUGGAUGUGGA--------- ....(((((.....(((.(((...((((.(((.....)))...)))))------))..)))................(((((((......)))))))..))))).......--------- ( -23.50) >DroSec_CAF1 20000 100 - 1 AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCU------GCGAUCUGU-----AAAAAAAAAUGUGUAUAAAAAGAUACACAGAUGUGGAUGUGGA--------- ...(((.(.......).)))......((((.(((.....(((((((..------..)))))))-----.........(((((((......)))))))...))).))))...--------- ( -24.10) >DroSim_CAF1 9985 100 - 1 AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCU------GCGAUCUGU-----AAAAAAAAAUGUGUAUAAAAAGAUACACAGAUGUGGAUGUGGA--------- ...(((.(.......).)))......((((.(((.....(((((((..------..)))))))-----.........(((((((......)))))))...))).))))...--------- ( -24.10) >DroEre_CAF1 11320 97 - 1 AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCU------GCGAUCUGU-----AA---AAAAUGUGUAUAAAAAGAUACACAGAUAUGGAUGUGGA--------- ....((((..........(((...((((.(((.....)))...)))))------)).((((((-----(.---....(((((((......)))))))..))))))))))).--------- ( -24.80) >DroYak_CAF1 11900 112 - 1 AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCUGCGUCUGCGAUCUGU-----AA---AAAAUGUGUAUAAAAAGAUACCCAGAUGUGGAUGUGGAUGUGGCUGC ..(((((((..((..(((...)))..)).....(((....)))..((..((((..(.(((((.-----..---.....(.((((......)))))))))))..))))..))))))))).. ( -33.60) >consensus AAGGCCACAAAGUAAGAGGCACUCGAGCAUGCACUGGGCACAGGUUCU______GCGAUCUGU_____AAAAAAAAAUGUGUAUAAAAAGAUACACAGAUGUGGAUGUGGA_________ ...(((.(.......).)))..((..((((.(((.....(((((((..........)))))))..............(((((((......)))))))...))).)))).))......... (-20.50 = -20.90 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:23 2006