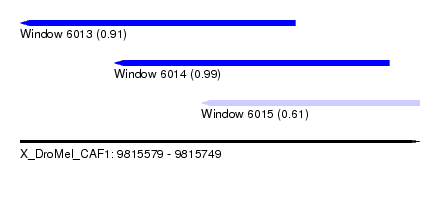
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,815,579 – 9,815,749 |
| Length | 170 |
| Max. P | 0.993582 |

| Location | 9,815,579 – 9,815,696 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9815579 117 - 22224390 GCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCACCUCAGCAAACAAAUUCCGAACUGGAGGCGAAUUUCCAGGAUAGCAGCGAUCCCGACUUGAAUCCUAUCAAC ((((......)))).((((((((......((((((...........))))))..............(((((((....)))))))))))))))..........((((......)))). ( -34.40) >DroSec_CAF1 10876 117 - 1 GCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCGGAACUGGAGGCAAAUUUCCAGGGUACCAGCGAUCCCGACUUGAAUCCUAUCAAC ((.((((.....)))).))..(((..((((((((.....((((((....)))...............((((((....)))))))))..))))))))..))).((((......)))). ( -37.60) >DroSim_CAF1 6667 117 - 1 GCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCGGAACUGGAGGCGAAUUUCCAGGAUACCAGCGAUCCCGACUUGAAUCCUAUCAAC ((.((((.....)))).))..(((..((((((((........(((....)))..............(((((((....)))))))....))))))))..))).((((......)))). ( -36.30) >DroEre_CAF1 6807 117 - 1 GCCGUGCUAUCGGCAUUGCUAUCGGUAACGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCCGAACUGGAGGCGAGAUUCCAGGAUACCAGCGAUCCCGACCUGAAUCCCAUCAAU ((((......)))).......((((...((((((........(((....)))..............((((((......))))))....))))))...))))................ ( -33.60) >DroYak_CAF1 8687 117 - 1 GCCGUGCUAUCGGCACUGCUAUCGCUAACGCUGGGCUAUCCCGCUCCACAGCAAACAAACUCCGAACUGGAGGCGAGUUUCCAGGAUACCAGCGAUCCCGACUUGAAUCCCAUCAAU ((.((((.....)))).))..(((....((((((..(((((.(((....)))((((...((((.....))))....))))...)))))))))))....))).((((......)))). ( -33.50) >consensus GCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCCGAACUGGAGGCGAAUUUCCAGGAUACCAGCGAUCCCGACUUGAAUCCUAUCAAC ((.((((.....)))).))..(((..((((((((........((......))..............(((((((....)))))))....))))))))..)))................ (-31.78 = -32.42 + 0.64)



| Location | 9,815,619 – 9,815,736 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.58 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9815619 117 - 22224390 AAUUAUGCAGUUCACCAGGAGCCGUUUAUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCACCUCAGCAAACAAAUUCCGAACUGGAGGCGAA .....(.((((((....((((..((((........(((((((.((((.....)))).))))))).....((((((...........))))))))))...)))))))))).)...... ( -34.80) >DroSec_CAF1 10916 117 - 1 AAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCGGAACUGGAGGCAAA .....(((......((((...(((((((((.....(((((((.((((.....)))).))))))).....(((((((......)))...)))).))))))..)))..))))..))).. ( -39.60) >DroSim_CAF1 6707 117 - 1 AAUAAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCGGAACUGGAGGCGAA .....(((......((((...(((((((((.....(((((((.((((.....)))).))))))).....(((((((......)))...)))).))))))..)))..))))..))).. ( -38.90) >DroEre_CAF1 6847 117 - 1 AAAUAUGCAGUUCACCAGGAGUCAUUUGUUGAUAUCGAUGGCCGUGCUAUCGGCAUUGCUAUCGGUAACGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCCGAACUGGAGGCGAG .....(.((((((....(((((..((((((((((((((((((.((((.....)))).))))))))))..((.(((....)))))...))))))))...))))))))))).)...... ( -43.00) >DroYak_CAF1 8727 117 - 1 AAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGCUAACGCUGGGCUAUCCCGCUCCACAGCAAACAAACUCCGAACUGGAGGCGAG .....(.((((((....((((..(((((((......((((((.((((.....)))).))))))((....))(((((......)).))).)))))))...)))))))))).)...... ( -40.50) >consensus AAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUCCCGCUCCUCAGCAAACAAAUUCCGAACUGGAGGCGAA .....(.((((((....(....)(((((((.....(((((((.((((.....)))).))))))).....(((((((......)))...)))).)))))))...)))))).)...... (-34.66 = -34.58 + -0.08)



| Location | 9,815,656 – 9,815,749 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9815656 93 - 22224390 ----------------------GCUUGCAGAAA-----CUAAUUAUGCAGUUCACCAGGAGCCGUUUAUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUC ----------------------(((((((....-----.......)))))....((((((...((.....))...(((((((.((((.....)))).)))))))...)).)))))).... ( -26.80) >DroSec_CAF1 10953 93 - 1 ----------------------GCUUGCAGAAA-----CUAAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUC ----------------------((((((.....-----.((((...((.((((.....)))).))..))))....(((((((.((((.....)))).))))))).....)).)))).... ( -27.00) >DroSim_CAF1 6744 93 - 1 ----------------------GCUUGCAGAAA-----CUAAUAAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAUCGCUGGGCUUUC ----------------------((((((.....-----.((((((.((.((((.....)))).))))))))....(((((((.((((.....)))).))))))).....)).)))).... ( -28.00) >DroEre_CAF1 6884 93 - 1 ----------------------GCUUGCAGAAA-----CUAAAUAUGCAGUUCACCAGGAGUCAUUUGUUGAUAUCGAUGGCCGUGCUAUCGGCAUUGCUAUCGGUAACGCUGGGCUUUC ----------------------(((((((....-----.......)))))....((((..((((.....))))(((((((((.((((.....)))).)))))))))....)))))).... ( -29.90) >DroYak_CAF1 8764 98 - 1 ----------------------GCUUGCAGAAAACUACUUAAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGCUAACGCUGGGCUAUC ----------------------.((((..((..(((.(........).)))))..))))((((.............((((((.((((.....)))).))))))((....))..))))... ( -27.30) >DroAna_CAF1 6706 116 - 1 CCAACCUCUCCUCUUCCUGUCACUUUCCAGAAGA----GCAGAGAUGCAGUUCCCCGUGCACCGAGUGUUGCUGUCGAUGGCCCUGUUAUCGGUGUCGCUGUCGCUGACGGCCGCCGCCC .....((((.((((((.((........)))))))----).))))..((((..((.((.(((........)))...))..))..))))...(((((..((((((...)))))))))))... ( -34.50) >consensus ______________________GCUUGCAGAAA_____CUAAUUAUGCAGUUCACCAGGAGCCGUUUGUUACUAUCGAUGGCCGUGCUAUCGGCACUGCUAUCGUUAACGCUGGGCUUUC ......................((((((...........((((...((.((((.....)))).))..))))....(((((((.((((.....)))).))))))).....)).)))).... (-20.42 = -20.12 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:17 2006