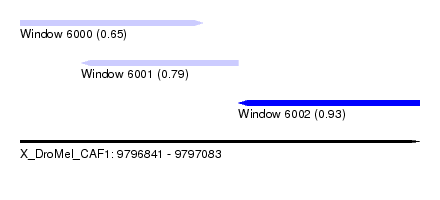
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,796,841 – 9,797,083 |
| Length | 242 |
| Max. P | 0.933928 |

| Location | 9,796,841 – 9,796,952 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -22.11 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9796841 111 + 22224390 ACACACUCACACACACACCACCC---CCCCUAUUUGACUCUACGCCCCUGUACUUUGGGGCCACCCUCUUUUAGCUGCCGUUUCAAUUUGGCCUUCUCAGAGGAACUGAGGUUG .....((((..............---.................(((((........)))))...(((((.......((((........))))......)))))...)))).... ( -25.62) >DroSec_CAF1 11542 104 + 1 ACAC----------ACACCACCCCCCCCCCUAUUUGACUCUACGCCCCUGUACUUUGGGGCCACCCUCUUUUAGCUGCCGUUUCAAUUUGGCCUUCUCAAGGGAACUGAGGUUG ....----------.......................(((...(((((........)))))..((((.....((..((((........)))))).....))))....))).... ( -23.70) >DroSim_CAF1 12679 101 + 1 ACAC----------ACACCACCC---CUCCUAUUUGACUCUACGCCCCUGUACUUUGGGGCCACCCUCUUUUAGCUGCCGUUUCAAUUUGGCCUUCUCAAGGGAACUGAGGUUG ....----------........(---(((..............(((((........)))))..((((.....((..((((........)))))).....))))....))))... ( -25.70) >consensus ACAC__________ACACCACCC___CCCCUAUUUGACUCUACGCCCCUGUACUUUGGGGCCACCCUCUUUUAGCUGCCGUUUCAAUUUGGCCUUCUCAAGGGAACUGAGGUUG .....................................(((...(((((........))))).....((((((((..((((........))))...)).))))))...))).... (-22.11 = -21.67 + -0.44)



| Location | 9,796,878 – 9,796,973 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.23 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.32 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9796878 95 - 22224390 AUGCUAAACAGCAAUGC--------C---AGC--------------CAACCUCAGUUCCUCUGAGAAGGCCAAAUUGAAACGGCAGCUAAAAGAGGGUGGCCCCAAAGUACAGGGGCGUA .((((....))))....--------(---(.(--------------(...(((((.....)))))...(((..........)))..........)).))(((((........)))))... ( -28.50) >DroPse_CAF1 22217 120 - 1 AUGCUAAACAGAGGAGGAGCUGGAGCUGGAGCUCUAGCUGGAGCAGGAGCAGGAGCACCCUCGAGCAGGCCAAAUUGAAACAGCAGCGUGGAGGGGGUGGUUGGGUGCUGGGUGGAGCCC ..(((..((........(((((((((....)))))))))....(((..((..((.(((((((.......(((..(((......)))..))).))))))).))..)).))).))..))).. ( -42.80) >DroSec_CAF1 11572 95 - 1 AUGCUAAACAGCAAUGC--------C---AGC--------------CAACCUCAGUUCCCUUGAGAAGGCCAAAUUGAAACGGCAGCUAAAAGAGGGUGGCCCCAAAGUACAGGGGCGUA .((((....))))((((--------(---.((--------------((.((((..(((......))).(((..........)))........)))).)))).((........))))))). ( -27.40) >DroSim_CAF1 12706 95 - 1 AUGCUAAACAGCAAUGC--------C---AGC--------------CAACCUCAGUUCCCUUGAGAAGGCCAAAUUGAAACGGCAGCUAAAAGAGGGUGGCCCCAAAGUACAGGGGCGUA .((((....))))((((--------(---.((--------------((.((((..(((......))).(((..........)))........)))).)))).((........))))))). ( -27.40) >consensus AUGCUAAACAGCAAUGC________C___AGC______________CAACCUCAGUUCCCUUGAGAAGGCCAAAUUGAAACGGCAGCUAAAAGAGGGUGGCCCCAAAGUACAGGGGCGUA .((((....))))............................................(((((.((...(((..........)))..))....)))))..(((((........)))))... (-17.45 = -17.32 + -0.13)



| Location | 9,796,973 – 9,797,083 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9796973 110 - 22224390 CGGAAAGGGGGGCUGGAAUAGC---CCCGGAAGAGGGGUGGUCCCAAAAAAAAAAAAAAAAACGAGGG-GACCAAAACAGUUUUGAAAACUGUAAUUAUUUUAAGGGAUUCUUU ........(((((((...))))---))).((((((...(((((((.....................))-)))))..((((((.....))))))...............)))))) ( -31.00) >DroSec_CAF1 11667 103 - 1 CGGAAAAGG-GGCCGGAAUAGCCGCCCCGAAAGAGGGGUGGUGCCAAAA----------AAUCGAGGGGGGCCAAAACAGUUUUGAAAACUGUAAUUAUUUUAAGGGAUUCUUU .((((....-((((((....((((((((......)))))))).))....----------..((....))))))...((((((.....))))))...............)))).. ( -32.80) >DroSim_CAF1 12801 102 - 1 CGGAAAGGG-GGCCGGAAUAGCCGCCCCGGAAGAGGGGUGGUGCCAAAA----------AACCGAGGG-GGCCAAAACAGUUUUGAAAACUGUAAUUAUUUUAAGGGAUUCUUU ...((((((-((((((....((((((((......)))))))).))....----------..((...))-))))...((((((.....))))))...............)))))) ( -34.60) >consensus CGGAAAGGG_GGCCGGAAUAGCCGCCCCGGAAGAGGGGUGGUGCCAAAA__________AAACGAGGG_GGCCAAAACAGUUUUGAAAACUGUAAUUAUUUUAAGGGAUUCUUU ....(((((...((((....((((((((......)))))))).))....................((....))...((((((.....))))))............))..))))) (-23.84 = -24.40 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:05 2006