
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,795,819 – 9,796,046 |
| Length | 227 |
| Max. P | 0.770148 |

| Location | 9,795,819 – 9,795,936 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.37 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9795819 117 + 22224390 AAUCCCAUCUGCCGGGCAUAUAUACGAAUGUGCAC--ACGUGUAUGUGGCAGAAUGGGAUGGAGAUAGAUGGAUGACCGGCAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACA .((((((((((((....(((((((((..((....)--))))))))))))))).))))))).................((.((((.......)))).)).......(((.......))). ( -33.80) >DroSec_CAF1 10505 118 + 1 AAUCCCAUCUGCCAGG-AUAUAUACGAAUGUGCACACACGCGUGUGUGGCAGAGUGGGAGGGAGAUAGAUGGAUGACCGGCAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACA ..((((((((((((..-...((((((..(((....)))..))))))))))))).)))))..................((.((((.......)))).)).......(((.......))). ( -34.50) >DroSim_CAF1 11638 118 + 1 AAUCCCAUCUGCCAGG-AUAUAUACGAAUGUGCACACACGCGUGUGUGGCAGAGUGGGAGGGAGAUAGAUGGAUGACCGGCAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACA ..((((((((((((..-...((((((..(((....)))..))))))))))))).)))))..................((.((((.......)))).)).......(((.......))). ( -34.50) >consensus AAUCCCAUCUGCCAGG_AUAUAUACGAAUGUGCACACACGCGUGUGUGGCAGAGUGGGAGGGAGAUAGAUGGAUGACCGGCAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACA ..((((((((((((......((((((..(((....)))..))))))))))))).)))))..................((.((((.......)))).)).......(((.......))). (-31.36 = -31.37 + 0.01)

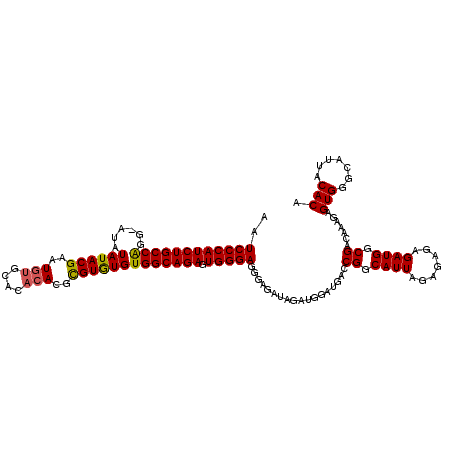

| Location | 9,795,897 – 9,796,006 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 99.39 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -31.39 |
| Energy contribution | -31.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9795897 109 + 22224390 CAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACAAAGGCCUUUUUGGCCGAUAAUGGAGCUUAUCUUGGCGGGGAUAAUGCCAGCAUAAAGCCGUAGGCAAAUA (((((....(.((((.(.((.....)).).)))).).....((((.....))))..)))))..((((((..(((((.......)))))((.....)).))))))..... ( -29.50) >DroSec_CAF1 10584 109 + 1 CAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACAAAGGCCUUUUUGGCCGAUAAUGGAGCUUAUCUUGGCGGGGAUAACGCCAGCAUAAAGCCGUAGGCAAAUA (((((....(.((((.(.((.....)).).)))).).....((((.....))))..)))))..((((((..(((((.......)))))((.....)).))))))..... ( -32.00) >DroSim_CAF1 11717 109 + 1 CAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACAAAGGCCUUUUUGGCCGAUAAUGGAGCUUAUCUUGGCGGGGAUAACGCCAGCAUAAAGCCGUAGGCAAAUA (((((....(.((((.(.((.....)).).)))).).....((((.....))))..)))))..((((((..(((((.......)))))((.....)).))))))..... ( -32.00) >consensus CAUUAGAGAGAGAUGGCGACAAAGAGUGGGCAUUACACAAAGGCCUUUUUGGCCGAUAAUGGAGCUUAUCUUGGCGGGGAUAACGCCAGCAUAAAGCCGUAGGCAAAUA (((((....(.((((.(.((.....)).).)))).).....((((.....))))..)))))..((((((..(((((.......)))))((.....)).))))))..... (-31.39 = -31.17 + -0.22)



| Location | 9,795,897 – 9,796,006 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 99.39 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9795897 109 - 22224390 UAUUUGCCUACGGCUUUAUGCUGGCAUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUUUGUGUAAUGCCCACUCUUUGUCGCCAUCUCUCUCUAAUG ..........((((.....(..(((((((..(.......(((((......)))))((((.....))))...)..)))))))..).....))))................ ( -24.50) >DroSec_CAF1 10584 109 - 1 UAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUUUGUGUAAUGCCCACUCUUUGUCGCCAUCUCUCUCUAAUG ...........(((.(((((((((((.......))))).(((((......)))))((((.....))))...)))))).)))............................ ( -26.10) >DroSim_CAF1 11717 109 - 1 UAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUUUGUGUAAUGCCCACUCUUUGUCGCCAUCUCUCUCUAAUG ...........(((.(((((((((((.......))))).(((((......)))))((((.....))))...)))))).)))............................ ( -26.10) >consensus UAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUUUGUGUAAUGCCCACUCUUUGUCGCCAUCUCUCUCUAAUG ...........(((.(((((((((((.......))))).(((((......)))))((((.....))))...)))))).)))............................ (-24.63 = -24.97 + 0.33)



| Location | 9,795,936 – 9,796,046 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 99.39 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -28.15 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9795936 110 - 22224390 AUAAAUAUGCGUGCUUGUUUGUUUCGGCCACCAAAUUGUGUAUUUGCCUACGGCUUUAUGCUGGCAUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUU ......(((.(.((((((((.....((((((......)))....((((...(((.....)))))))........))).)))))))).))))....((((.....)))).. ( -28.40) >DroSec_CAF1 10623 110 - 1 AUAAAUAUGCGUGCUUGUUUGUUUCGGCCACCAAAUUGUGUAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUU ......(((.(.((((((((((...((((..(((((.....))))).....))))....))(((((.......))))))))))))).))))....((((.....)))).. ( -29.10) >DroSim_CAF1 11756 110 - 1 AUAAAUAUGCGUGCUUGUUUGUUUCGGCCACCAAAUUGUGUAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUU ......(((.(.((((((((((...((((..(((((.....))))).....))))....))(((((.......))))))))))))).))))....((((.....)))).. ( -29.10) >consensus AUAAAUAUGCGUGCUUGUUUGUUUCGGCCACCAAAUUGUGUAUUUGCCUACGGCUUUAUGCUGGCGUUAUCCCCGCCAAGAUAAGCUCCAUUAUCGGCCAAAAAGGCCUU ......(((.(.((((((((.....((((((......)))....((((...(((.....)))))))........))).)))))))).))))....((((.....)))).. (-28.15 = -27.93 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:00 2006