
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,794,143 – 9,794,456 |
| Length | 313 |
| Max. P | 0.987910 |

| Location | 9,794,143 – 9,794,245 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794143 102 - 22224390 AACACAAAGAGCAGGGGAAUUGCUGUAGCCAAUUUAUUGGUUAAUUGCCAUUUCAAUCAUUUUGAUUGUGGAAUUCAGCUAAGCUCUUGGACCACAAGACCA ...........((((((....(((((((((((....)))))))....((((.((((.....))))..))))....))))....))))))............. ( -25.30) >DroSec_CAF1 8865 102 - 1 AACACAAAGAGCAGGGGAAUUGCUGGAGCCAAUUUAUUGGUUAAUUGCCAUUUCAAUCAUUUUGAUUGUGGAAUUCAGCUAAGCUCUUCGGCCACAAGCCCA ......((((((....(((((((....).)))))).((((((((((.((((.((((.....))))..)))))))).)))))))))))).(((.....))).. ( -25.60) >DroSim_CAF1 9999 102 - 1 AACACAAAGAGCAGGGGAAUUGCUGGAGCCAAUUUAUUGGUUAAUUGCCAUUUCAAUCAUUUUGAUUGUGGAAUUCAGCUAAGCUCUUGGGCCACAAGCCCA ......((((((....(((((((....).)))))).((((((((((.((((.((((.....))))..)))))))).))))))))))))((((.....)))). ( -30.10) >consensus AACACAAAGAGCAGGGGAAUUGCUGGAGCCAAUUUAUUGGUUAAUUGCCAUUUCAAUCAUUUUGAUUGUGGAAUUCAGCUAAGCUCUUGGGCCACAAGCCCA ......((((((....(((((((....).)))))).((((((((((.((((.((((.....))))..)))))))).))))))))))))((((.....)))). (-25.17 = -25.83 + 0.67)

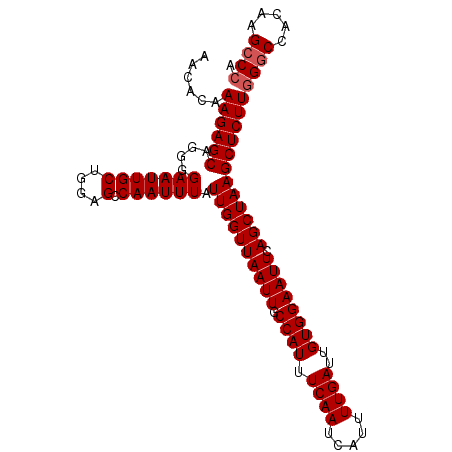

| Location | 9,794,245 – 9,794,340 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 98.60 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794245 95 + 22224390 UGCCAGUUGGGCGGUGAGGAAUGCCCACGAGGCCUUCAUAGAUUCAAAUAUGCAAUCAAAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUA .....((.(((((........)))))))..((((.....((((((......((.......))(((((((...)))))))...))))))))))... ( -25.80) >DroSec_CAF1 8967 95 + 1 UGCCAGUUGGGCGGCGAGGAAUGCCCACGAGGCCUUCAUAGACUCAAAUAUGCAAUCAAAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUA .....((.(((((........)))))))..(((((((.(((.((((((.((((.........))))))))))......))).)))...))))... ( -27.60) >DroSim_CAF1 10101 95 + 1 UGCCAGUUGGGCGGCGAGGAAUGCCCACGAGGCCUUCAUAGAUUCAAAUAUGCAAUCAAAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUA .(((.....)))(((...(((.(((.....))).))).(((((((......((.......))(((((((...)))))))...))))))))))... ( -26.90) >consensus UGCCAGUUGGGCGGCGAGGAAUGCCCACGAGGCCUUCAUAGAUUCAAAUAUGCAAUCAAAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUA .....((.(((((........)))))))..(((((((.(((.((((((.((((.........))))))))))......))).)))...))))... (-26.35 = -26.13 + -0.22)


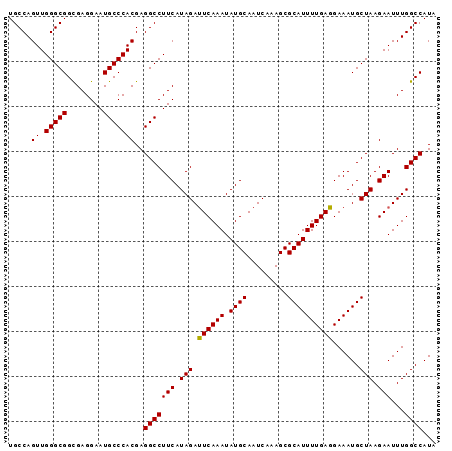
| Location | 9,794,245 – 9,794,340 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 98.60 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794245 95 - 22224390 UAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUUUGAUUGCAUAUUUGAAUCUAUGAAGGCCUCGUGGGCAUUCCUCACCGCCCAACUGGCA (((((.(((((.....((((((.....))))))((.......))..)))))...)))))...(((...(((((..........)))))...))). ( -24.00) >DroSec_CAF1 8967 95 - 1 UAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUUUGAUUGCAUAUUUGAGUCUAUGAAGGCCUCGUGGGCAUUCCUCGCCGCCCAACUGGCA ...((((((((.....((((((.....))))))((.......))..))))).((((((((.....)))))))).......)))(((.....))). ( -25.90) >DroSim_CAF1 10101 95 - 1 UAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUUUGAUUGCAUAUUUGAAUCUAUGAAGGCCUCGUGGGCAUUCCUCGCCGCCCAACUGGCA (((((.(((((.....((((((.....))))))((.......))..)))))...)))))...(((...(((((..........)))))...))). ( -24.00) >consensus UAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUUUGAUUGCAUAUUUGAAUCUAUGAAGGCCUCGUGGGCAUUCCUCGCCGCCCAACUGGCA (((((.(((((.....((((((.....))))))((.......))..)))))...)))))...(((...(((((..........)))))...))). (-24.00 = -24.00 + 0.00)

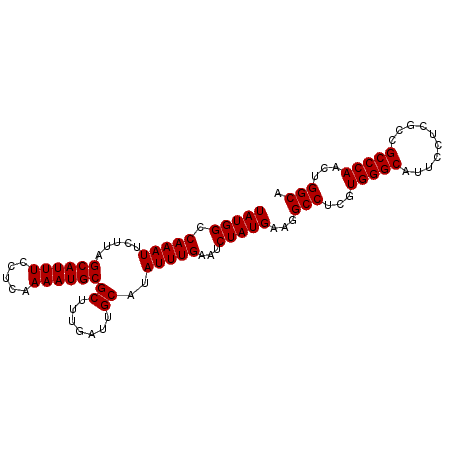
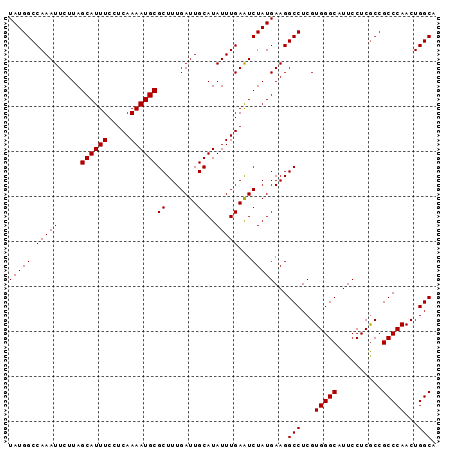
| Location | 9,794,303 – 9,794,418 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 98.26 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -43.89 |
| Energy contribution | -44.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794303 115 + 22224390 AAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUAAUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCUGCUACUUCAGUUCCUCCUGACCGCUUUUCGGCGGC ....(((((((...))))))).........((((.........))))..(((((.((((..((((.((((.....)))).)))).....)))).))))).(((((....))))). ( -39.60) >DroSec_CAF1 9025 115 + 1 AAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUAAUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGC ....(((((((...))))))).........((((.........))))..(((((.((((..(((((((((.....))))))))).....)))).))))).(((((....))))). ( -46.00) >DroSim_CAF1 10159 115 + 1 AAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUAAUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGC ....(((((((...))))))).........((((.........))))..(((((.((((..(((((((((.....))))))))).....)))).))))).(((((....))))). ( -46.00) >consensus AAGCGCAUUUUGAGGAAAUGCUAAGAAUUUGGCCAUAAUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGC ....(((((((...))))))).........((((.........))))..(((((.((((..(((((((((.....))))))))).....)))).))))).(((((....))))). (-43.89 = -44.00 + 0.11)



| Location | 9,794,303 – 9,794,418 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 98.26 |
| Mean single sequence MFE | -45.23 |
| Consensus MFE | -44.50 |
| Energy contribution | -44.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794303 115 - 22224390 GCCGCCGAAAAGCGGUCAGGAGGAACUGAAGUAGCAGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAUUAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUU (((((......)))))(((((.((.((...(((((.((((.....)))).))))))).)).)))))..((((.........)))).........((((((.....)))))).... ( -39.90) >DroSec_CAF1 9025 115 - 1 GCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAUUAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUU ((((((....))))))(((((.((.((...(((((.((((.....)))).))))))).)).)))))..((((.........)))).........((((((.....)))))).... ( -47.90) >DroSim_CAF1 10159 115 - 1 GCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAUUAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUU ((((((....))))))(((((.((.((...(((((.((((.....)))).))))))).)).)))))..((((.........)))).........((((((.....)))))).... ( -47.90) >consensus GCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAUUAUGGCCAAAUUCUUAGCAUUUCCUCAAAAUGCGCUU ((((((....))))))(((((.((.((...(((((.((((.....)))).))))))).)).)))))..((((.........)))).........((((((.....)))))).... (-44.50 = -44.83 + 0.33)

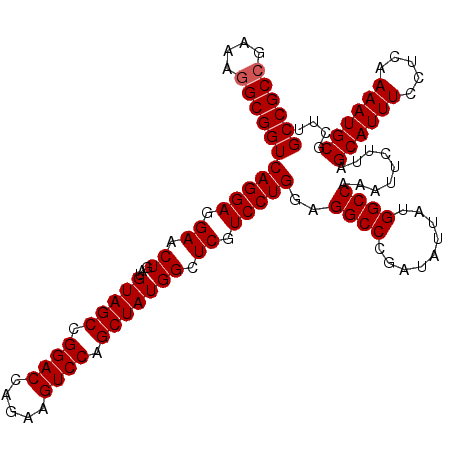

| Location | 9,794,340 – 9,794,456 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 97.70 |
| Mean single sequence MFE | -46.13 |
| Consensus MFE | -45.39 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794340 116 + 22224390 AUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCUGCUACUUCAGUUCCUCCUGACCGCUUUUCGGCGGCUUGCGGUUGCCCAAAAUCGAAGGCAAAGCGAUGGGGAA .(((((.((((.(((((.((((..((((.((((.....)))).)))).....)))).)))))..((.((((.((((((.....)))))).)))).)).))))....)))))..... ( -42.80) >DroSec_CAF1 9062 116 + 1 AUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGCUUGCGGUUGCCCAAAAUCGAAGGCAAAGCGAUGGGAAA .(((((((((..(((((.((((..(((((((((.....))))))))).....)))).)))))...(((....))))))))((..(((((..........))))).))))))..... ( -47.50) >DroSim_CAF1 10196 116 + 1 AUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGCUUGCGGUUGCCCAAAAUCGAAGGCAAAGCGAUGGGGAA ........(((((((((.((((..(((((((((.....))))))))).....)))).))))).(((((....))))).((((..(((((..........))))).)))).)))).. ( -48.10) >consensus AUAUCGGGCCUCCAGGACGAGCCAUAGCUGGACUUCUGGUCCGGCUACAUCAGUUCCUCCUGACCGCCUUUCGGCGGCUUGCGGUUGCCCAAAAUCGAAGGCAAAGCGAUGGGGAA .(((((((((..(((((.((((..(((((((((.....))))))))).....)))).)))))...(((....))))))))((..(((((..........))))).))))))..... (-45.39 = -45.50 + 0.11)



| Location | 9,794,340 – 9,794,456 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.70 |
| Mean single sequence MFE | -47.03 |
| Consensus MFE | -46.30 |
| Energy contribution | -46.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794340 116 - 22224390 UUCCCCAUCGCUUUGCCUUCGAUUUUGGGCAACCGCAAGCCGCCGAAAAGCGGUCAGGAGGAACUGAAGUAGCAGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAU ......((((....((((((......((....))....(((((......))))).((((.((.((...(((((.((((.....)))).))))))).)).)))))))))).)))).. ( -41.70) >DroSec_CAF1 9062 116 - 1 UUUCCCAUCGCUUUGCCUUCGAUUUUGGGCAACCGCAAGCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAU ......((((....((((((......((....))....((((((....)))))).((((.((.((...(((((.((((.....)))).))))))).)).)))))))))).)))).. ( -49.70) >DroSim_CAF1 10196 116 - 1 UUCCCCAUCGCUUUGCCUUCGAUUUUGGGCAACCGCAAGCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAU ......((((....((((((......((....))....((((((....)))))).((((.((.((...(((((.((((.....)))).))))))).)).)))))))))).)))).. ( -49.70) >consensus UUCCCCAUCGCUUUGCCUUCGAUUUUGGGCAACCGCAAGCCGCCGAAAGGCGGUCAGGAGGAACUGAUGUAGCCGGACCAGAAGUCCAGCUAUGGCUCGUCCUGGAGGCCCGAUAU ......((((....((((((......((....))....((((((....)))))).((((.((.((...(((((.((((.....)))).))))))).)).)))))))))).)))).. (-46.30 = -46.63 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:57 2006