| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,794,024 – 9,794,132 |
| Length | 108 |
| Max. P | 0.886900 |

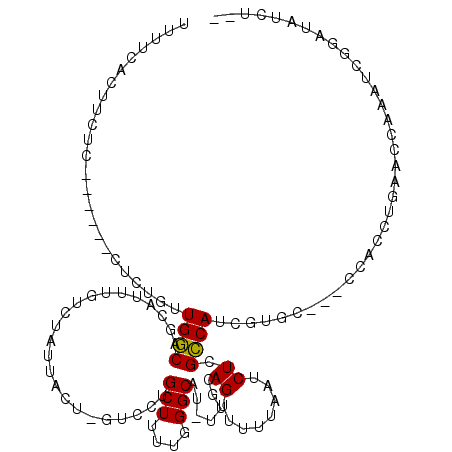
| Location | 9,794,024 – 9,794,132 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.70 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -7.26 |
| Energy contribution | -6.78 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794024 108 + 22224390 UUUUCACUUCUC------CUCUGUUGGCAGUAUUUGUCUAUUACUUGUCCUGCUUUUGGGCAU-UUGCAGUUUUUAAUCUAGCCAUAGUGC---CCACCUGAAUCAAAUCGGAUAUCU-- ..........((------(......(((((.((..((.....))..)).)))))..(((((((-(.((((........)).))...)))))---))).............))).....-- ( -22.30) >DroPse_CAF1 19246 107 + 1 UUUCCGCUUUUCGUUCGCUUUUGUUGCCACCAAA-----AGCACU-----CGCUUUUGGGCAU-UUGCAGUGUUUAAUCUUGGCAACGAGAAUGCCAGCUG--CCAAAUGAGAUUUCUAC .....(((...(((((....((((((((((((((-----(((...-----.))))))))((..-..))............))))))))))))))..)))..--................. ( -30.90) >DroSec_CAF1 8744 109 + 1 UUUUCACUUCUC------CUCUGUUGGCAGCAUUUGUCUAUUACUCGUCCUGCUUUUGGGCAUUUUGCAGUUUUUAAUCUCGCCAUUGUGC---CCACCUGAACCAAAUCGGAAAUCU-- ..........((------(....(((((((.((..((.....))..)).))))...(((((((..(((((........)).).))..))))---))).......)))...))).....-- ( -21.30) >DroSim_CAF1 9878 109 + 1 UUUUCACUUCUC------CUCUGUUGGCAGCAUUUGUCUAUUACUAGUCCUGCUUUUGGGCAUUUUGCAGUUUUUAAUCUCGCCAUUGUGC---CCACCUGAACCAAAUCGGAAAUCU-- ..........((------(....((((.((((...(.(((....))).).))))..(((((((..(((((........)).).))..))))---)))......))))...))).....-- ( -22.30) >DroPer_CAF1 16021 107 + 1 UUUCCGCUUUUCGUUCGCUUUUGUUGCCACCAAA-----AGCACU-----CGCUUUUGGGCAU-UUGCAGUGUUUAAUCUUGGCAACGAGAAUGCCAGCUG--CCAAAUGAGAUUUCUAC .....(((...(((((....((((((((((((((-----(((...-----.))))))))((..-..))............))))))))))))))..)))..--................. ( -30.90) >consensus UUUUCACUUCUC______CUCUGUUGGCAGCAUUUGUCUAUUACU_GUCCUGCUUUUGGGCAU_UUGCAGUUUUUAAUCUCGCCAUCGUGC___CCACCUGAACCAAAUCGGAUAUCU__ ........................((((.......................(((....))).......((........)).))))................................... ( -7.26 = -6.78 + -0.48)

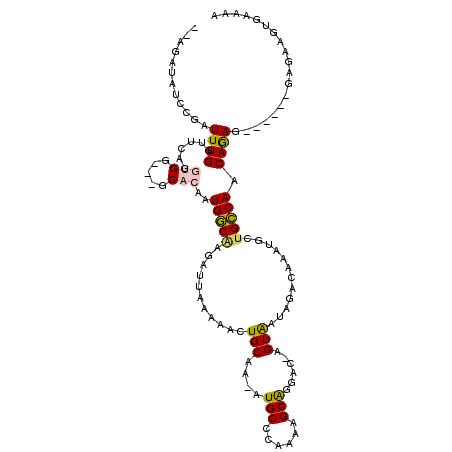
| Location | 9,794,024 – 9,794,132 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.70 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -7.50 |
| Energy contribution | -7.14 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9794024 108 - 22224390 --AGAUAUCCGAUUUGAUUCAGGUGG---GCACUAUGGCUAGAUUAAAAACUGCAA-AUGCCCAAAAGCAGGACAAGUAAUAGACAAAUACUGCCAACAGAG------GAGAAGUGAAAA --.....(((..((((.......(((---(((.....((.((........))))..-.))))))...((((.....((.....)).....))))...)))))------)).......... ( -19.50) >DroPse_CAF1 19246 107 - 1 GUAGAAAUCUCAUUUGG--CAGCUGGCAUUCUCGUUGCCAAGAUUAAACACUGCAA-AUGCCCAAAAGCG-----AGUGCU-----UUUGGUGGCAACAAAAGCGAACGAAAAGCGGAAA .....(((((....(((--((((.((....)).)))))))))))).....((((..-....((((((((.-----...)))-----)))))((....))..............))))... ( -30.00) >DroSec_CAF1 8744 109 - 1 --AGAUUUCCGAUUUGGUUCAGGUGG---GCACAAUGGCGAGAUUAAAAACUGCAAAAUGCCCAAAAGCAGGACGAGUAAUAGACAAAUGCUGCCAACAGAG------GAGAAGUGAAAA --...(((((...(((((.....(((---(((.....((.((........))))....))))))..((((.(.(........).)...)))))))))....)------))))........ ( -24.10) >DroSim_CAF1 9878 109 - 1 --AGAUUUCCGAUUUGGUUCAGGUGG---GCACAAUGGCGAGAUUAAAAACUGCAAAAUGCCCAAAAGCAGGACUAGUAAUAGACAAAUGCUGCCAACAGAG------GAGAAGUGAAAA --...(((((...(((((.....(((---(((.....((.((........))))....))))))..((((.(.(((....))).)...)))))))))....)------))))........ ( -27.90) >DroPer_CAF1 16021 107 - 1 GUAGAAAUCUCAUUUGG--CAGCUGGCAUUCUCGUUGCCAAGAUUAAACACUGCAA-AUGCCCAAAAGCG-----AGUGCU-----UUUGGUGGCAACAAAAGCGAACGAAAAGCGGAAA .....(((((....(((--((((.((....)).)))))))))))).....((((..-....((((((((.-----...)))-----)))))((....))..............))))... ( -30.00) >consensus __AGAUAUCCGAUUUGGUUCAGGUGG___GCACAAUGGCAAGAUUAAAAACUGCAA_AUGCCCAAAAGCAGGAC_AGUAAUAGACAAAUGCUGCCAACAGAG______GAGAAGUGAAAA ............((((......(((.....)))..(((((...........(((....(((......)))......)))............))))).))))................... ( -7.50 = -7.14 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:51 2006