
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,793,135 – 9,793,415 |
| Length | 280 |
| Max. P | 0.989219 |

| Location | 9,793,135 – 9,793,255 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.09 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793135 120 - 22224390 AUGGUGAAGAACUAGAACAAAAAAGGGCUUCAAGCUGGGAGCGAUUUAGUUUGAAUACUCAUAGAGAUUUCAGACUAAAUCAAUAGAUCAAGAUCUAUAGUUAGCCCUACUGUUUGAUUU .((((.....))))(((((....((((((...(((((.....(((((((((((((..(((...)))..)))))))))))))..(((((....))))))))))))))))..)))))..... ( -36.40) >DroSec_CAF1 7876 114 - 1 AUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAG-----UCUGAUUUGGUGUGAAUACUCAUUGAAAACUCAGACUAAAAAAUAAGAUACAAGUCUGUAGUUAGCCUUACUUUUUGAAU- .((((.....))))...((((((((((((.(.((-----(((((((..(((........)))..))...)))))))........((((....))))...)..))))))..))))))...- ( -26.20) >DroSim_CAF1 9014 119 - 1 AUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAGCUGGGUCUGAUUUGGUGUGAAUACUCAUUGAAAACUCAGACUAAAAAAUAAGAUACAAAUCUGUAGUUAGCCUUACUUUUUGAAU- .((((.....))))...((((((((((((...((((((((((((((..(((........)))..))...)))))))........((((....)))).)))))))))))..))))))...- ( -28.60) >consensus AUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAGCUGGGUCUGAUUUGGUGUGAAUACUCAUUGAAAACUCAGACUAAAAAAUAAGAUACAAAUCUGUAGUUAGCCUUACUUUUUGAAU_ .((((.....))))...((((((((((((.(.............((((((.(((...............))).)))))).....((((....))))...)..))))))..)))))).... (-19.64 = -19.09 + -0.55)


| Location | 9,793,215 – 9,793,335 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -33.80 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793215 120 + 22224390 UCCCAGCUUGAAGCCCUUUUUUGUUCUAGUUCUUCACCAUAGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAA ...(((((.((.((((((...((...........))....)))))).)).)))))...((((((((((((((...((((((....)))))).......)))))...)))))))))..... ( -37.30) >DroSec_CAF1 7955 115 + 1 A-----CUUGAAGCCCUUUCUUGUUCUAGUUCUUCACCAUAGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAA .-----..(((((..((..........))..))))).....((((((......))))))(((((((((((((...((((((....)))))).......)))))...))))))))...... ( -33.80) >DroSim_CAF1 9093 120 + 1 ACCCAGCUUGAAGCCCUUUCUUGUUCUAGUUCUUCACCAUAGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAA ...(((((.((.((((((...((...........))....)))))).)).)))))...((((((((((((((...((((((....)))))).......)))))...)))))))))..... ( -37.30) >consensus ACCCAGCUUGAAGCCCUUUCUUGUUCUAGUUCUUCACCAUAGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAA ........(((((..((..........))..))))).....((((((......))))))(((((((((((((...((((((....)))))).......)))))...))))))))...... (-33.80 = -33.80 + 0.00)


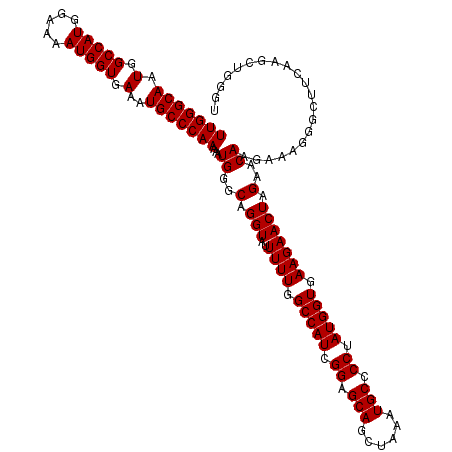
| Location | 9,793,215 – 9,793,335 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -37.71 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793215 120 - 22224390 UUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCUAUGGUGAAGAACUAGAACAAAAAAGGGCUUCAAGCUGGGA (((((((.(.(((((.....))))).)..)))))))..((((.((((....)))).)))).....(((((....((((...((((.....))))..........))))....)))))... ( -38.52) >DroSec_CAF1 7955 115 - 1 UUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCUAUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAG-----U (((((((.(.(((((.....))))).)..)))))))...((..(.(((..((((.(((((.((.(((......))).)).))))).))))))).)..)).(((.....)))...-----. ( -36.10) >DroSim_CAF1 9093 120 - 1 UUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCUAUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAGCUGGGU (((((((.(.(((((.....))))).)..)))))))..((((.((((....)))).)))).....(((((....((((...((((.....))))..........))))....)))))... ( -38.52) >consensus UUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCUAUGGUGAAGAACUAGAACAAGAAAGGGCUUCAAGCUGGGU (((((((.(.(((((.....))))).)..)))))))...((..(.(((..((((.(((((.((.(((......))).)).))))).))))))).)..))..................... (-36.00 = -36.00 + 0.00)


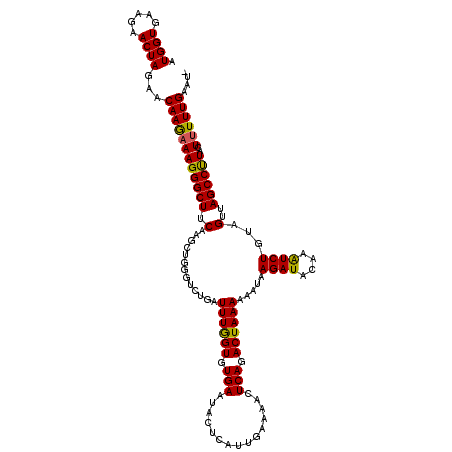
| Location | 9,793,255 – 9,793,375 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -38.89 |
| Energy contribution | -38.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793255 120 + 22224390 AGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCCGAACAAAGGCCGCAUUAAGUGUCC ..((((((((((.(((..((((((((((((((...((((((....)))))).......)))))...)))))))))..................(((.......))).))))))))))))) ( -40.40) >DroSec_CAF1 7990 120 + 1 AGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUGUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCC ..((((((((((.(((..((((((((((((((...((((((....)))))).......)))))...))))))))).......((.((((((....)))))).))...))))))))))))) ( -40.90) >DroSim_CAF1 9133 120 + 1 AGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCC ..((((((((((.(((..((((((((((((((...((((((....)))))).......)))))...)))))))))..................(((.......))).))))))))))))) ( -37.80) >consensus AGGGGCAUUUAGCUGCUCCGAUGGCCAAAAAUACCUGCCCAUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCC ..((((((((((.(((..((((((((((((((...((((((....)))))).......)))))...)))))))))..................(((.......))).))))))))))))) (-38.89 = -38.67 + -0.22)

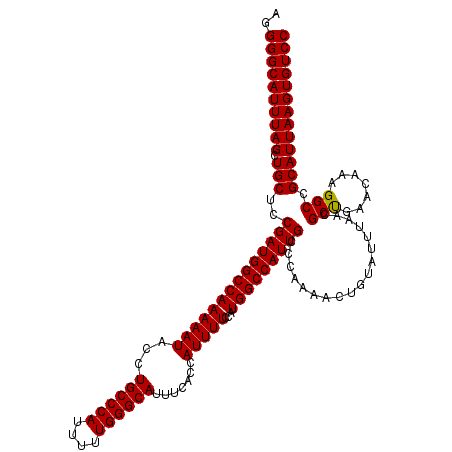
| Location | 9,793,255 – 9,793,375 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -39.25 |
| Energy contribution | -39.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793255 120 - 22224390 GGACACUUAAUGCGGCCUUUGUUCGGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCU ((.((.(((.(((((((.((((((((..((......))..)))))))).))))((((..((..(((...((((((....))))))..)))..))..))))....)))..))).))))... ( -38.50) >DroSec_CAF1 7990 120 - 1 GGACACUUAAUGCGGCCUUUGUUCAGCGUUAAACACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCU ((.((.(((.(((((((.((((((((..((......))..)))))))).))))((((..((..(((...((((((....))))))..)))..))..))))....)))..))).))))... ( -39.30) >DroSim_CAF1 9133 120 - 1 GGACACUUAAUGCGGCCUUUGUUCAGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCU ((.((.(((.(((((((.((((((((..((......))..)))))))).))))((((..((..(((...((((((....))))))..)))..))..))))....)))..))).))))... ( -39.30) >consensus GGACACUUAAUGCGGCCUUUGUUCAGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAUGGGCAGGUAUUUUUGGCCAUCGGAGCAGCUAAAUGCCCCU ((.((.(((.(((((((.((((((((..((......))..)))))))).))))((((..((..(((...((((((....))))))..)))..))..))))....)))..))).))))... (-39.25 = -39.03 + -0.22)



| Location | 9,793,295 – 9,793,415 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793295 120 + 22224390 AUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCCGAACAAAGGCCGCAUUAAGUGUCCUAGCAGCCGCAGGCAGGAUGAAGGAUAAAGGAGUCCUGCU ..(((((((((.....((((.....))))....)))))))))((((.......(((.......)))(((.....)))...........))))(((((((.............))))))). ( -34.32) >DroSec_CAF1 8030 120 + 1 AUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUGUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCCUGGCAGCCGCAGGCAGGAUGAAGGAUAAAGGAGUCCUGCU ..(((((((((.....((((.....))))....)))))))))(((((......((((.....(((.(((.....))).)))..)))))))))(((((((.............))))))). ( -40.82) >DroSim_CAF1 9173 120 + 1 AUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCCUGGCAGCCGCAGGCAGGAUGAAGGAUAAAGGAGUCCUGCU ..(((((((((.....((((.....))))....)))))))))((((.......((((.....(((.(((.....))).)))..)))).))))(((((((.............))))))). ( -37.82) >consensus AUUUUUGGGCAUUUCACCAUUUUCCAUGGCCAUUGCCCAAAACUGUAUUUAACGCUGAACAAAGGCCGCAUUAAGUGUCCUGGCAGCCGCAGGCAGGAUGAAGGAUAAAGGAGUCCUGCU ..(((((((((.....((((.....))))....)))))))))((((.......((((.....(((.(((.....))).)))..)))).))))(((((((.............))))))). (-35.72 = -36.05 + 0.33)


| Location | 9,793,295 – 9,793,415 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.09 |
| Consensus MFE | -36.98 |
| Energy contribution | -36.76 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9793295 120 - 22224390 AGCAGGACUCCUUUAUCCUUCAUCCUGCCUGCGGCUGCUAGGACACUUAAUGCGGCCUUUGUUCGGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAU .((((((...............))))))....((((((.((....))....)))))).....................(((((((((.(.(((((.....))))).)..))))))))).. ( -37.56) >DroSec_CAF1 8030 120 - 1 AGCAGGACUCCUUUAUCCUUCAUCCUGCCUGCGGCUGCCAGGACACUUAAUGCGGCCUUUGUUCAGCGUUAAACACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAU .((((((...............))))))(((.((((((.((....))....))))))..((((........)))))))(((((((((.(.(((((.....))))).)..))))))))).. ( -37.56) >DroSim_CAF1 9173 120 - 1 AGCAGGACUCCUUUAUCCUUCAUCCUGCCUGCGGCUGCCAGGACACUUAAUGCGGCCUUUGUUCAGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAU .((((((...............))))))....((((((.((....))....)))))).....................(((((((((.(.(((((.....))))).)..))))))))).. ( -36.16) >consensus AGCAGGACUCCUUUAUCCUUCAUCCUGCCUGCGGCUGCCAGGACACUUAAUGCGGCCUUUGUUCAGCGUUAAAUACAGUUUUGGGCAAUGGCCAUGGAAAAUGGUGAAAUGCCCAAAAAU .((((((...............))))))(((.((((((.((....))....))))))..((((........)))))))(((((((((.(.(((((.....))))).)..))))))))).. (-36.98 = -36.76 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:46 2006