
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,791,255 – 9,791,507 |
| Length | 252 |
| Max. P | 0.842408 |

| Location | 9,791,255 – 9,791,375 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -38.22 |
| Energy contribution | -39.33 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9791255 120 + 22224390 UUAAGUUUCUCGGCUCUUUUUUCCGGGCCGAGAACAAUGUCGGCAUAGUUGGUCGAUGGGCGGAGCGAGCAGAAAUUGCCCGGGGGCAAGAAGGCCAACUAAUGGCAAGGACUUGAGUCC ....(((.((((((((........)))))))))))..(((((...(((((((((.....((.......)).....(((((....)))))...))))))))).))))).((((....)))) ( -48.10) >DroSec_CAF1 5987 117 + 1 UUCAGUUUCUAGGUUCUUUUUUCCGGGCCGAGAACAAUGUCGGC--AGUUGGUCGAUGGGCGGAGCCAGUAGAAAUUGGCCGG-GGCAAGAAGGCCAACUAAUGGCAAGGACUUGAGUCC ....(((.((.(((((........))))).)))))..(((((..--((((((((.....((...((((((....))))))...-.)).....))))))))..))))).((((....)))) ( -39.70) >DroSim_CAF1 7132 117 + 1 UUAAGUUUCUCGGUUCUUUUUUCCGGGCCGAGAACAAUGUCGGC--AGUUGGUCGAUGGGCGGAGCCAGUAGAAAUUGGCCGG-GGCAAGAAGGCCAACUAAUGGCAAGGACUUGAGUCC ....(((.((((((((........)))))))))))..(((((..--((((((((.....((...((((((....))))))...-.)).....))))))))..))))).((((....)))) ( -45.60) >consensus UUAAGUUUCUCGGUUCUUUUUUCCGGGCCGAGAACAAUGUCGGC__AGUUGGUCGAUGGGCGGAGCCAGUAGAAAUUGGCCGG_GGCAAGAAGGCCAACUAAUGGCAAGGACUUGAGUCC ....(((.((((((((........)))))))))))..(((((....((((((((.....((...((((((....)))))).....)).....))))))))..))))).((((....)))) (-38.22 = -39.33 + 1.11)



| Location | 9,791,375 – 9,791,493 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9791375 118 + 22224390 GCAAAUUCCAGCUAAAUCUAUACCUACAAUACUGCAUUAUCUCGCAUUUGAGCCAAAAACGCAACCCGACAUCGAAUUGGCAAUCGAUUCCAGCUGC--ACUUGCCUUCAAUCCACCAAG ((((....(((((.((((........(((...(((........))).))).(((((...((...........))..)))))....))))..))))).--..))))............... ( -19.10) >DroSec_CAF1 6104 120 + 1 GCAAAUUCCAGCUAAAUCUAUACCUACAAUACUGCAUUAUCUCGCAUUUGAGCCAAAAACGCAACCCGACAUCGAAUUGGCAAUCGAUUCCAGCUGCACACUUGCCUUCAAUCCACCAAU ((((....(((((.((((........(((...(((........))).))).(((((...((...........))..)))))....))))..))))).....))))............... ( -18.90) >DroSim_CAF1 7249 120 + 1 GCAAAUUCCAGCUAAAUCUAUACCUACAAUACUGCAUUAUCUCGCAUUUGAGCCAAAAACGCAACCCGACAUCGAAUUGGCAAUCGAUUCCAGCUGCACACUUGCCUUCAACCGACCAAU ((((....(((((.((((........(((...(((........))).))).(((((...((...........))..)))))....))))..))))).....))))............... ( -18.90) >consensus GCAAAUUCCAGCUAAAUCUAUACCUACAAUACUGCAUUAUCUCGCAUUUGAGCCAAAAACGCAACCCGACAUCGAAUUGGCAAUCGAUUCCAGCUGCACACUUGCCUUCAAUCCACCAAU ((((....(((((.((((........(((...(((........))).))).(((((...((...........))..)))))....))))..))))).....))))............... (-18.90 = -18.90 + 0.00)

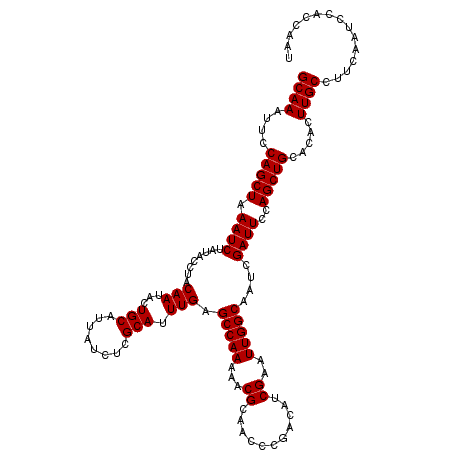
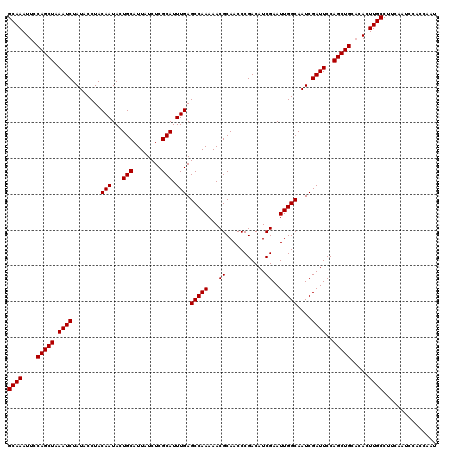
| Location | 9,791,375 – 9,791,493 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -34.37 |
| Energy contribution | -34.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9791375 118 - 22224390 CUUGGUGGAUUGAAGGCAAGU--GCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAGAUAAUGCAGUAUUGUAGGUAUAGAUUUAGCUGGAAUUUGC (((.(.....).)))((((((--.((((((.((((...((((((((((....))))))((((((((.((......)).))).))))).........))))..))))))))))..)))))) ( -35.00) >DroSec_CAF1 6104 120 - 1 AUUGGUGGAUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAGAUAAUGCAGUAUUGUAGGUAUAGAUUUAGCUGGAAUUUGC ((((.((.(((..((.((((..(((((((.(((((((........))))).)).)))))))..)))).))..))).))...))))((((.(((.(((.((......)).))).))))))) ( -34.60) >DroSim_CAF1 7249 120 - 1 AUUGGUCGGUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAGAUAAUGCAGUAUUGUAGGUAUAGAUUUAGCUGGAAUUUGC ((((.(((.((((.(.((((..(((((((.(((((((........))))).)).)))))))..)))).)))))...)))..))))((((.(((.(((.((......)).))).))))))) ( -35.20) >consensus AUUGGUGGAUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAGAUAAUGCAGUAUUGUAGGUAUAGAUUUAGCUGGAAUUUGC ...............((((((...((((((.((((...((((((((((....))))))((((((((.((......)).))).))))).........))))..))))))))))..)))))) (-34.37 = -34.37 + 0.00)

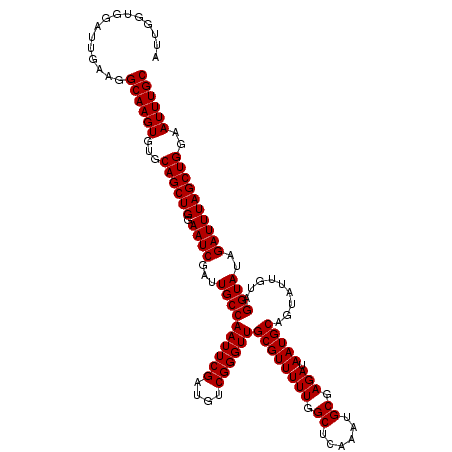
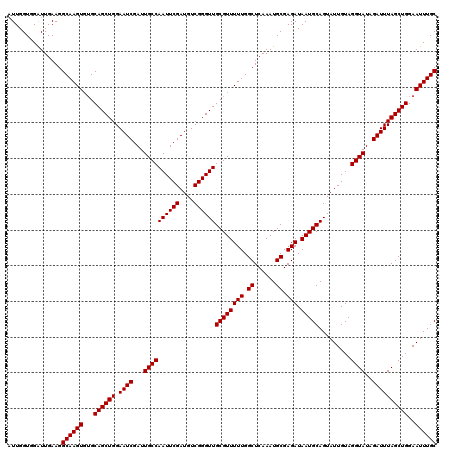
| Location | 9,791,415 – 9,791,507 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9791415 92 - 22224390 AUAUAAUAUUACAUCUUGGUGGAUUGAAGGCAAGU--GCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAG ...........(((....)))..((((.(.(((((--((((((.(((((((........))))).)).)))))))..)))).)))))....... ( -24.00) >DroSec_CAF1 6144 94 - 1 AUAUAAUAUUAUAUAUUGGUGGAUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAG ...................((.(((..((.((((..(((((((.(((((((........))))).)).)))))))..)))).))..))).)).. ( -26.70) >DroSim_CAF1 7289 94 - 1 AUAUAAUAUUAUAUAUUGGUCGGUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAG ...................(((.((((.(.((((..(((((((.(((((((........))))).)).)))))))..)))).)))))...))). ( -27.70) >consensus AUAUAAUAUUAUAUAUUGGUGGAUUGAAGGCAAGUGUGCAGCUGGAAUCGAUUGCCAAUUCGAUGUCGGGUUGCGUUUUUGGCUCAAAUGCGAG ..................((...((((.(.((((..(((((((.(((((((........))))).)).)))))))..)))).)))))..))... (-24.00 = -24.33 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:40 2006