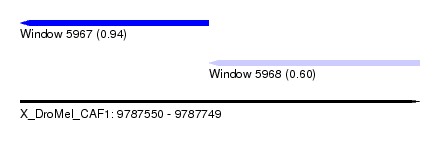
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,787,550 – 9,787,749 |
| Length | 199 |
| Max. P | 0.937969 |

| Location | 9,787,550 – 9,787,644 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -29.07 |
| Energy contribution | -28.63 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.937969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
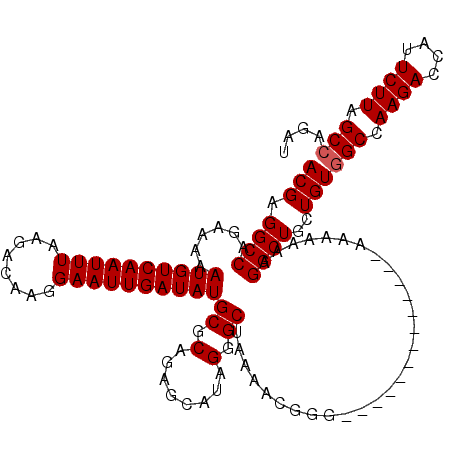
>X_DroMel_CAF1 9787550 94 - 22224390 UAGGCCAUGCCCUGCCAGCAGAUGGCCAUAAGCGGGGGUGAAU-----------CCCUGGGAUGUCCUUCGACCCUUCGACUGGCAUAUGCACCCUCUUACUCCC--------------- ..((((((...(((....)))))))))......(((((((.((-----------((...))))(((..((((....))))..))).....)))))))........--------------- ( -32.40) >DroEre_CAF1 83197 105 - 1 UAGGCCAUGCCCUGCCAGCAGCUGGCCAUAAUCGGGGGUGAAAUAAAUACGAUAUCCAGGGAUGUCCUUCGACCCUUCGACUGGCAUAUGCACCUUCCUACUCUC--------------- ..(((((.((..((....)))))))))......(((((((..........((((((....))))))..((((....))))..........)))))))........--------------- ( -29.50) >DroYak_CAF1 74482 120 - 1 UAGGCCGUGCCCUGCCAGCAGCUGGCCAUAAUCGGGGGUGAAAUAAAUACGACAUCCAGGGAUGUCCUUCGACCCUUCGACUGGCAUAUGCACCCUCCUACUCCCUCUCUCCCUCUCUCU ..(((((.((..((....)))))))))......(((((((..........((((((....))))))..((((....))))..........)))))))....................... ( -33.50) >consensus UAGGCCAUGCCCUGCCAGCAGCUGGCCAUAAUCGGGGGUGAAAUAAAUACGA_AUCCAGGGAUGUCCUUCGACCCUUCGACUGGCAUAUGCACCCUCCUACUCCC_______________ ..(((((....(((....))).)))))......(((((((...............(....)(((((..((((....))))..)))))...)))))))....................... (-29.07 = -28.63 + -0.44)


| Location | 9,787,644 – 9,787,749 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9787644 105 - 22224390 ACGAGGCCAGAAAAAUGUCAAUUUAAGACAAGGAAUUGAUAUGCGCAGAGCAUAGGGCUGAAACG---------------UAAAAAAGGCUGCUGUGGCCAAGACCAUUCUUAGCGAGAU .((.((((......((((((((((........))))))))))..(((((((.....((......)---------------).......))).))))))))((((....))))..)).... ( -25.50) >DroEre_CAF1 83302 109 - 1 ACGAGGCCAGAAAAAUGUCAAUUUAAGACAAGGAAUUGAUAUGCGCAGAGCAUAGGGCUAAAACGGGG-----------GAAAAAAAGGCUGCUGUGGCCAAGACCAUUCUUUGCCAGAU ....((((......((((((((((........))))))))))((.....))....))))...((((.(-----------(.........)).))))(((.((((....)))).))).... ( -27.30) >DroYak_CAF1 74602 120 - 1 ACGAGGCCGGAAAAAUGUCAAUUUAAGACAAGGAAUUGAUAUGCGCAGAGCAUAGGGCUAAAACGGGCCAAAGAAACAAAAAAAAAAGGCUGCUGUGGCCAAGAGCAUUCUUAGCCAGAU (((.((((......((((((((((........))))))))))((.....))....((((......))))..................))))..)))(((.(((......))).))).... ( -29.50) >consensus ACGAGGCCAGAAAAAUGUCAAUUUAAGACAAGGAAUUGAUAUGCGCAGAGCAUAGGGCUAAAACGGG_____________AAAAAAAGGCUGCUGUGGCCAAGACCAUUCUUAGCCAGAU (((.((((......((((((((((........))))))))))((.(........).)).............................))))..)))(((.((((....)))).))).... (-22.70 = -23.03 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:35 2006