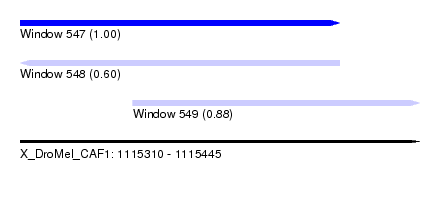
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,115,310 – 1,115,445 |
| Length | 135 |
| Max. P | 0.999958 |

| Location | 1,115,310 – 1,115,418 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -34.90 |
| Energy contribution | -35.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 1.04 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1115310 108 + 22224390 GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------UGAGGACGUUGCCGGGA-UCUCU ..(((((.....(((....)))..)))))................(((((((((......((((((((...)))))))).((((..-------...))))))))))))).-..... ( -37.20) >DroSec_CAF1 11922 108 + 1 GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGCCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGACGUUGCCGGGA-UCUCU ..(((((.....(((....)))..)))))................((((.((((......((((((((...)))))))).((((..-------...)))))))).)))).-..... ( -30.10) >DroSim_CAF1 12529 108 + 1 GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGACGUUGCCGGGA-UCUCU ..(((((.....(((....)))..)))))................(((((((((......((((((((...)))))))).((((..-------...))))))))))))).-..... ( -37.20) >DroEre_CAF1 13095 116 + 1 GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUUUGUCCUACGAGGACGUCGUCGGGAAUCUUU ..(((((.....(((....)))..)))))................((((((.((...(((((((((((...)))))).)))))....((((.....)))))).))))))....... ( -32.90) >DroYak_CAF1 12100 107 + 1 GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGGCGUCGUCGGGA-UCUU- ..(((((.....(((....)))..)))))................((((((.((......((((((((...)))))))).((((..-------...)))))).)))))).-....- ( -29.80) >consensus GCUGUAAUAUAAUCCUAAUGGAAUUUACACUCAAACUUUACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU_______CGAGGACGUUGCCGGGA_UCUCU ..(((((.....(((....)))..)))))................(((((((((......((((((((...)))))))).((((((........)))))))))))))))....... (-34.90 = -35.18 + 0.28)

| Location | 1,115,310 – 1,115,418 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1115310 108 - 22224390 AGAGA-UCCCGGCAACGUCCUCA-------AAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGCCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC ...((-(((.((((((((((...-------..))))..(((((((...))))))).......)))))).)).))).......((((((((((..(((....)))))))))).))). ( -33.80) >DroSec_CAF1 11922 108 - 1 AGAGA-UCCCGGCAACGUCCUCG-------AAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGGCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC .....-(((.(....)((((...-------..))))...)))...((((((.(((.((((....))))))))))))).....((((((((((..(((....)))))))))).))). ( -32.40) >DroSim_CAF1 12529 108 - 1 AGAGA-UCCCGGCAACGUCCUCG-------AAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGCCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC ...((-(((.((((((((((...-------..))))..(((((((...))))))).......)))))).)).))).......((((((((((..(((....)))))))))).))). ( -33.80) >DroEre_CAF1 13095 116 - 1 AAAGAUUCCCGACGACGUCCUCGUAGGACAAAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGCCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC ...(((.((.(.((((((((.....))))...((....(((((((...)))))))..))...)))).).)).))).......((((((((((..(((....)))))))))).))). ( -29.40) >DroYak_CAF1 12100 107 - 1 -AAGA-UCCCGACGACGCCCUCG-------AAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGCCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC -..((-(((..(.((.((.((((-------((..(((((((((((...))))))).(((..........))).))))....)))))).))....))..)..))))).......... ( -24.40) >consensus AGAGA_UCCCGGCAACGUCCUCG_______AAGGACAAAGGACACGCAGUGUCCUCGCCAUUGUUGCCAGGCAUUGUAAAGUUUGAGUGUAAAUUCCAUUAGGAUUAUAUUACAGC ..........(((...(((((..........)))))..(((((((...))))))).)))...((((.(((((........)))))(((((((..(((....)))))))))).)))) (-25.76 = -26.36 + 0.60)

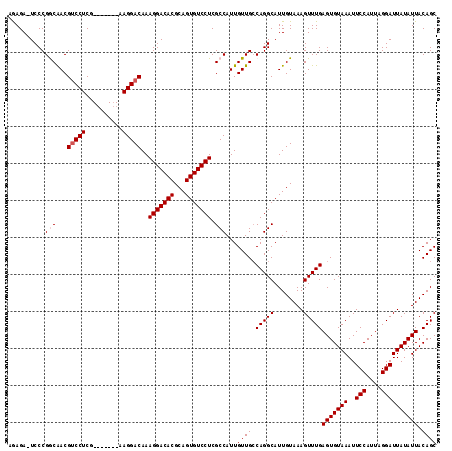
| Location | 1,115,348 – 1,115,445 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1115348 97 + 22224390 UACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------UGAGGACGUUGCCGGGA-UCUCUCUGC-UGGCGAAAGGAAGCAAUAGCGCC------------ .......(((((((((......((((((((...)))))))).((((..-------...))))))))))))).-......(((-(..(....)..))))........------------ ( -39.80) >DroSec_CAF1 11960 97 + 1 UACAAUGCCUGCCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGACGUUGCCGGGA-UCUCUCUUC-UGGCGAAAGGAAGCAAAAGCGCC------------ .....(((((((((....)))).))).)).(((...((((((((((..-------...))))..((((((((-......)))-))))))))))).)))........------------ ( -35.00) >DroSim_CAF1 12567 97 + 1 UACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGACGUUGCCGGGA-UCUCUCUUC-UGGCGAAAGGAAGCAAUAGCGCC------------ .......(((((((((......((((((((...)))))))).((((..-------...))))))))))))).-.........-.((((...(....).....))))------------ ( -37.10) >DroEre_CAF1 13133 118 + 1 UACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUUUGUCCUACGAGGACGUCGUCGGGAAUCUUUCUGCUGUGCGAAAGGAAGCAAUAUAGGCAGCAAUGGAGAG .......((((((.((...(((((((((((...)))))).)))))....((((.....)))))).))))))..((((((((((.(((........))).....)))))....))))). ( -40.00) >DroYak_CAF1 12138 100 + 1 UACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU-------CGAGGGCGUCGUCGGGA-UCUU--CGCCUGGCGAAAGGAAGCAAUUGAGGCAACC-------- .....((((((....)...((((((((..((((((((((((.......-------.))))))).)).)))..-))))--))))..((........)).....)))))...-------- ( -38.50) >consensus UACAAUGCCUGGCAACAAUGGCGAGGACACUGCGUGUCCUUUGUCCUU_______CGAGGACGUUGCCGGGA_UCUCUCUGC_UGGCGAAAGGAAGCAAUAGCGCC____________ .......(((((((((......((((((((...)))))))).((((((........)))))))))))))))..............((........))..................... (-32.30 = -32.58 + 0.28)

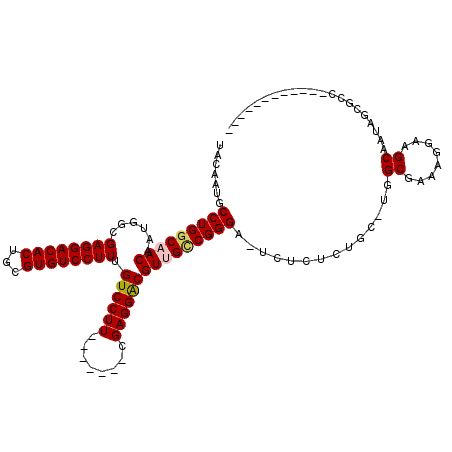
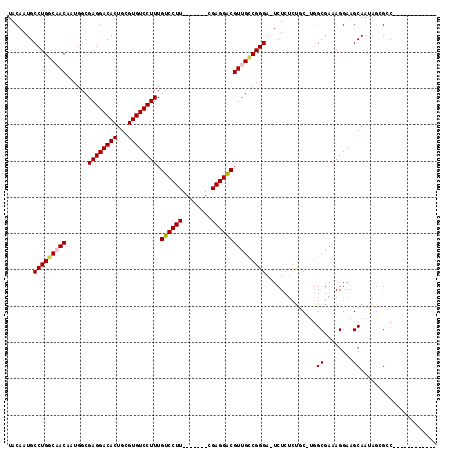
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:40 2006