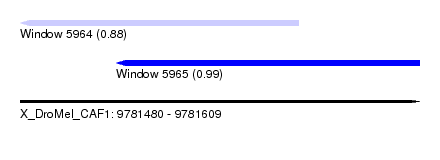
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,781,480 – 9,781,609 |
| Length | 129 |
| Max. P | 0.993397 |

| Location | 9,781,480 – 9,781,570 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9781480 90 - 22224390 UAGUUUAGAUAGUCUAUAGUCGAAAGGAAGUGGAAGUGGCACCACAGUGGUGAAGCGAUCGGGUACGUAAACAAAUUUCUAUCUACAAAU----- .....((((((((((((..((....))..))))).((..((((.....))))..))......................))))))).....----- ( -22.10) >DroSec_CAF1 77762 87 - 1 UAGUUUAGAUAGUCUAUAGUCGAAAGGAAGUGGAGGUGGCACCACAGUGGUGAAGCGAUCGGAGAGGUAAACAAAUUUCUAUCUAAA-------- ...((((((((((((((..((....))..))))).((..((((.....))))..))......................)))))))))-------- ( -23.20) >DroYak_CAF1 68568 90 - 1 UAGUUUAGAUAGUCUAUAGUCGAAAGGAAAUGGAGGUGGCACCACAGUGGUGAAGCGAUCGGAGACCUGAACAAAUUUCUGU-----AAUAUUUG ..((((((....(((((..((....))..))))).((..((((.....))))..))....(....)))))))..........-----........ ( -21.70) >consensus UAGUUUAGAUAGUCUAUAGUCGAAAGGAAGUGGAGGUGGCACCACAGUGGUGAAGCGAUCGGAGACGUAAACAAAUUUCUAUCUA_AAAU_____ .....((((((((((((..((....))..))))).((..((((.....))))..))......................))))))).......... (-19.24 = -19.80 + 0.56)

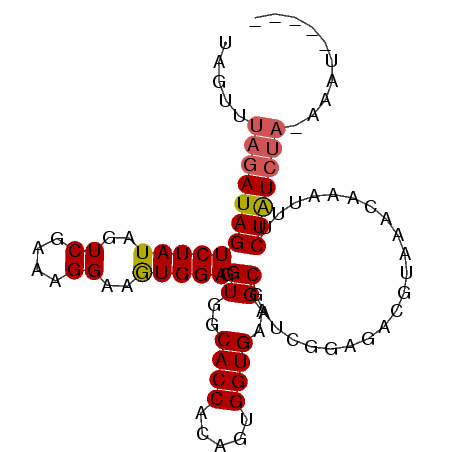
| Location | 9,781,511 – 9,781,609 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -12.42 |
| Energy contribution | -11.42 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9781511 98 - 22224390 AAGCGAAAGCAAUUCAGGCGUGCCAGGAUAUUUAGACUUUAGUU-UAGAUAGUC--UAUAGUCGAAA-----------GGAAGUGGAAGUGGCACCACAGUGGUGAAGCGAU--- ..((....)).......((((((((...(((((((((....)))-)))))).((--(((..((....-----------))..)))))..))))))(((....)))..))...--- ( -32.30) >DroSec_CAF1 77790 98 - 1 AAGCGAAAGCAAUUCAGGCGUGCCAGGAUAUUUAGACUUUAGUU-UAGAUAGUC--UAUAGUCGAAA-----------GGAAGUGGAGGUGGCACCACAGUGGUGAAGCGAU--- ..((....)).......((((((((...(((((((((....)))-)))))).((--(((..((....-----------))..)))))..))))))(((....)))..))...--- ( -32.30) >DroEre_CAF1 76067 101 - 1 CAGAGAAAGCAAUUCAGGCGUGCCAGGAUAUUUAGACUUUAGUU-UAGAUAGUC--CAUAGUCGAAA-----------GGAAGUGGAGGUGGCACCACAGUGGUGAGGCGAUCGG ....((..((.........((((((...(((((((((....)))-)))))).((--(((..((....-----------))..)))))..))))))(((....)))..))..)).. ( -31.10) >DroYak_CAF1 68599 98 - 1 AGGAGACAGCAAUUCAGGCGUGCCAGGAUAUUUAGACUUUAGUU-UAGAUAGUC--UAUAGUCGAAA-----------GGAAAUGGAGGUGGCACCACAGUGGUGAAGCGAU--- .(....).((.........((((((...(((((((((....)))-)))))).((--(((..((....-----------))..)))))..))))))(((....)))..))...--- ( -29.00) >DroAna_CAF1 52568 103 - 1 -----AAACCAAUUGAGGCAUGCCAGGACACUUUGACUUCAGUUGUGGACUGCCAGGACAGUCGAAAAUGGUGGCCAAGGAUGUG----UGAAAUGGCGGUUGUUAAGUUAA--- -----...(((.((.(.((((.(..((.((((((((((...(((.(((....))).)))))))))....)))).))..).)))).----).)).)))...............--- ( -27.20) >consensus AAGAGAAAGCAAUUCAGGCGUGCCAGGAUAUUUAGACUUUAGUU_UAGAUAGUC__UAUAGUCGAAA___________GGAAGUGGAGGUGGCACCACAGUGGUGAAGCGAU___ .................((((((((...(((((.(((....)))...(((..........))).................)))))....))))))(((....)))..))...... (-12.42 = -11.42 + -1.00)

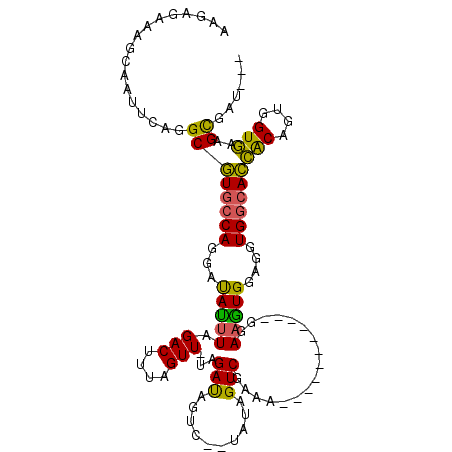

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:32 2006