
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,780,419 – 9,780,544 |
| Length | 125 |
| Max. P | 0.993230 |

| Location | 9,780,419 – 9,780,519 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.34 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9780419 100 - 22224390 CAGAUGCCAUAUCCUUUGUGGCGAUACUCUAGGGUAUCGAUCGAGCAUAUUGGCACGCCAAGCUGCAAG-GACACAUAAGAUCUUUUGGGAAAUUAAGUAC---- ((((.((((((.....))))))((((((....))))))((((..(((..((((....))))..)))..(-....)....)))).)))).............---- ( -27.90) >DroSec_CAF1 76736 101 - 1 CAGAUGCCAUAUCCUUUGUGGCGAUACUCUAGGGUAUCGAUCGAGUAUAUUGGCAAGCCAA--UGCAAA-AGCACUUAAAACCGUUGGGAGAAUUAGUUUA-AUA ..(((((((((.....))))))((((((....)))))).)))((((..(((((....))))--)((...-.))))))....((....))............-... ( -25.50) >DroEre_CAF1 75042 96 - 1 CGGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCGAGCGUAUC---GAGCCAC--UGCACA-GGCAGUUAAAA--GUCAGGAAAAUA-AAUAAAUAA .((..((((((.....))))))((((((....))))))..((((.....))---)).))((--(((...-.))))).....--............-......... ( -26.90) >DroYak_CAF1 67512 93 - 1 CAGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCAAGCAUAUUGGCAAGGCAC--UGCAAAAGGUUUUCGAAA--GCUUGGAAAAAG--------AG .....((((((.....))))))((((((....))))))..((((((...((.(.(((((.(--(.....)))))))).)).--))))))......--------.. ( -26.10) >consensus CAGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCGAGCAUAUUGGCAAGCCAA__UGCAAA_GGCACUUAAAA__GUUUGGAAAAUU_A_UAA__AA ..(((((((((.....))))))((((((....)))))).)))..(((...(((....)))...)))....................................... (-19.65 = -20.02 + 0.38)

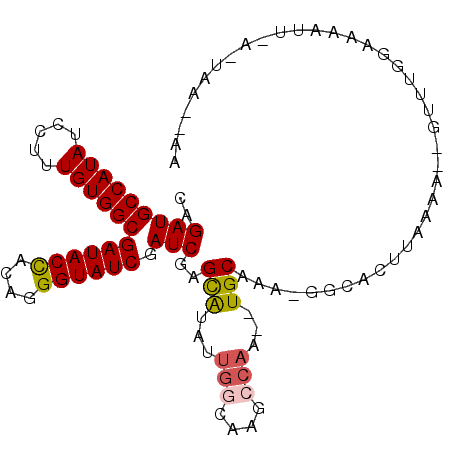

| Location | 9,780,451 – 9,780,544 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -20.33 |
| Energy contribution | -22.57 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
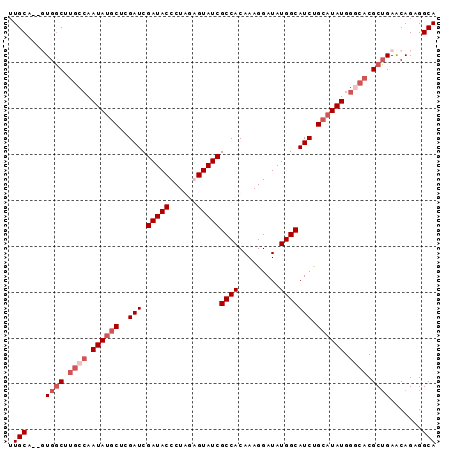
>X_DroMel_CAF1 9780451 93 + 22224390 UUGCAGCUUGGCGUGCCAAUAUGCUCGAUCGAUACCCUAGAGUAUCGCCACAAAGGAUAUGGCAUCUGCAUAUGGGCACGCUGAACAGAGGCA .(((..((..(((((((.((((((..(((.(((((......)))))((((.........))))))).)))))).)))))))..)...)..))) ( -36.60) >DroSec_CAF1 76771 91 + 1 UUGCA--UUGGCUUGCCAAUAUACUCGAUCGAUACCCUAGAGUAUCGCCACAAAGGAUAUGGCAUCUGCAUAUGGGCACGCUGAACAGAGGCA .(((.--(..((.((((.((((.(..(((.(((((......)))))((((.........))))))).).)))).)))).))..)......))) ( -23.00) >DroEre_CAF1 75075 88 + 1 GUGCA--GUGGCUC---GAUACGCUCGAUCGAUACCCUGUGGUAUCGCCACAAAGGAUAUGGCAUCCGCAUAUGGGAACGCUGAGCAGAGGCA ((((.--(((((((---((.....))))..((((((....)))))))))))...((((.....))))))))........(((.......))). ( -29.70) >DroYak_CAF1 67539 91 + 1 UUGCA--GUGCCUUGCCAAUAUGCUUGAUCGAUACCCUGUGGUAUCGCCACAAAGGAUAUGGCAUCUGCAUAUGGCCACGCGGAACAGAGGCA .....--.((((((((((.(((.(((...(((((((....))))))).....))).))))))).(((((..........)))))...)))))) ( -31.90) >consensus UUGCA__GUGGCUUGCCAAUAUGCUCGAUCGAUACCCUAGAGUAUCGCCACAAAGGAUAUGGCAUCUGCAUAUGGGCACGCUGAACAGAGGCA .(((....((((.((((.((((((..(((.(((((......)))))((((.........))))))).)))))).)))).)))).......))) (-20.33 = -22.57 + 2.25)


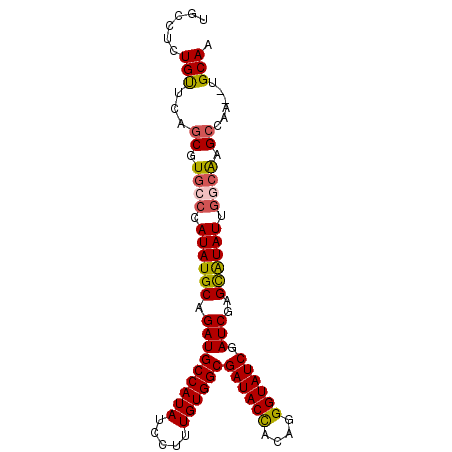
| Location | 9,780,451 – 9,780,544 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -28.73 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9780451 93 - 22224390 UGCCUCUGUUCAGCGUGCCCAUAUGCAGAUGCCAUAUCCUUUGUGGCGAUACUCUAGGGUAUCGAUCGAGCAUAUUGGCACGCCAAGCUGCAA (((....(((..(((((((.((((((.(((((((((.....))))))((((((....)))))).)))..)))))).)))))))..))).))). ( -43.60) >DroSec_CAF1 76771 91 - 1 UGCCUCUGUUCAGCGUGCCCAUAUGCAGAUGCCAUAUCCUUUGUGGCGAUACUCUAGGGUAUCGAUCGAGUAUAUUGGCAAGCCAA--UGCAA ......(((...((.((((.((((((.(((((((((.....))))))((((((....)))))).)))..)))))).)))).))...--.))). ( -33.40) >DroEre_CAF1 75075 88 - 1 UGCCUCUGCUCAGCGUUCCCAUAUGCGGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCGAGCGUAUC---GAGCCAC--UGCAC (((....((((.(((((((((((...(((((...)))))..)))))(((((((....)))))))...))))))...---))))...--.))). ( -32.20) >DroYak_CAF1 67539 91 - 1 UGCCUCUGUUCCGCGUGGCCAUAUGCAGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCAAGCAUAUUGGCAAGGCAC--UGCAA ......(((...((.(.(((((((((....((((((.....))))))((((((....))))))......))))).)))).).))..--.))). ( -34.10) >consensus UGCCUCUGUUCAGCGUGCCCAUAUGCAGAUGCCAUAUCCUUUGUGGCGAUACCACAGGGUAUCGAUCGAGCAUAUUGGCAAGCCAA__UGCAA ......(((...((.((((.((((((.(((((((((.....))))))((((((....)))))).)))..)))))).)))).))......))). (-28.73 = -28.98 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:30 2006