
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,772,576 – 9,772,673 |
| Length | 97 |
| Max. P | 0.996650 |

| Location | 9,772,576 – 9,772,673 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
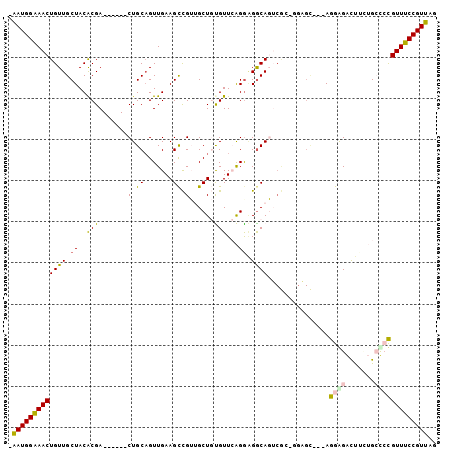
>X_DroMel_CAF1 9772576 97 + 22224390 -AAUGGAAACUGUUGCUACACGA------CUGCAGUUGAAGCCGUUGCUGUGUUCAGAAGGCAGUCGCUGGAGCAGGAGGGGAUUUUUGCUCCGUUUCCGUUAG -(((((((((..........(((------((((..(((((..((....))..)))))...)))))))..((((((((((....))))))))))))))))))).. ( -39.80) >DroPse_CAF1 75258 98 + 1 GGAUGGAAACUGUUGCUACACGACGACGACUGCAGUUGAAGUUGGGGCU-CGUUGGGGGGGCAUGCAG-CGUGU---AGAAGGCCA-AGGCAUGUUUCCGUUGG .(((((((((.....((((((..((((.......))))..((((.((((-(.......)))).).)))-)))))---))...((..-..))..))))))))).. ( -33.90) >DroSec_CAF1 69073 97 + 1 -AAUGGAAACUGUUGCUACACGA------CUGCCGUUGAAGCCGUUGCUGUGUUCAGGCGACAGUCGCGGGAGCAGGAGGAGAUUUCUGCUCCGUUUCCGUUAG -(((((((((........(.(((------(((.((((..(((((....)).)))..)))).)))))).)((((((((((....))))))))))))))))))).. ( -42.10) >DroEre_CAF1 67608 85 + 1 -AAUGGAAACUGUUGCUACGCGA------CUGCAGUUGAAGCCGUUGCUGUGUUCAGGAGGCAGU------------AGGGGACUUCUGCCCCGUUUCCGUUAG -(((((((((((..((.((((((------(.((.......)).))))).)))).))((.(((((.------------((....)).)))))))))))))))).. ( -35.90) >DroYak_CAF1 59536 97 + 1 -AAUGGGAACUGUUGCUACGCGA------CUGCAGUUGAAGCCGUUGCUGUGUUCAGGAGGCAGUCGCUGAAGCAGGAGGGGAUUUCUGCUCCGUUUCCGUUAG -(((((((((...((((.(((((------((((..(((((..((....))..)))))...)))))))).).))))((((.((....)).))))))))))))).. ( -42.30) >DroPer_CAF1 79879 98 + 1 GGAUGGAAACUGUUGCUACACGACGAUGACUGCAGUUGAAGUUGGGGCU-CGUUGGGGGGGCAUGCAG-CGUGU---AGAAGGCCA-AGGCAUGUUUCCGUUGG .(((((((((...((((...((((((.(.((.(((......))).))))-)))))....(((.(((..-...))---)....))).-.)))).))))))))).. ( -32.40) >consensus _AAUGGAAACUGUUGCUACACGA______CUGCAGUUGAAGCCGUUGCUGUGUUCAGGAGGCAGUCGC_GGAGC___AGGAGACUUCUGCCCCGUUUCCGUUAG .(((((((((((((.((..(((.......(.((.......)).)......)))...)).))))...............((((.......))))))))))))).. (-15.42 = -15.04 + -0.39)


| Location | 9,772,576 – 9,772,673 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.22 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9772576 97 - 22224390 CUAACGGAAACGGAGCAAAAAUCCCCUCCUGCUCCAGCGACUGCCUUCUGAACACAGCAACGGCUUCAACUGCAG------UCGUGUAGCAACAGUUUCCAUU- .....((((((((((((............)))))).((((((((....(((....(((....))))))...))))------)))).........))))))...- ( -33.50) >DroPse_CAF1 75258 98 - 1 CCAACGGAAACAUGCCU-UGGCCUUCU---ACACG-CUGCAUGCCCCCCCAACG-AGCCCCAACUUCAACUGCAGUCGUCGUCGUGUAGCAACAGUUUCCAUCC .....((((((..((..-..)).....---....(-(((((((........(((-(((..((........))..))..))))))))))))....)))))).... ( -22.90) >DroSec_CAF1 69073 97 - 1 CUAACGGAAACGGAGCAGAAAUCUCCUCCUGCUCCCGCGACUGUCGCCUGAACACAGCAACGGCUUCAACGGCAG------UCGUGUAGCAACAGUUUCCAUU- .....(((((((((((((..........)))))))(((((((((((..(((....(((....)))))).))))))------)))))........))))))...- ( -41.60) >DroEre_CAF1 67608 85 - 1 CUAACGGAAACGGGGCAGAAGUCCCCU------------ACUGCCUCCUGAACACAGCAACGGCUUCAACUGCAG------UCGCGUAGCAACAGUUUCCAUU- .....(((((((((((((.((....))------------.))))))).........((..(((((.(....).))------)))....))....))))))...- ( -26.90) >DroYak_CAF1 59536 97 - 1 CUAACGGAAACGGAGCAGAAAUCCCCUCCUGCUUCAGCGACUGCCUCCUGAACACAGCAACGGCUUCAACUGCAG------UCGCGUAGCAACAGUUCCCAUU- .....((.(((((((..(....)..))))((((...((((((((....(((....(((....))))))...))))------))))..))))...))).))...- ( -32.00) >DroPer_CAF1 79879 98 - 1 CCAACGGAAACAUGCCU-UGGCCUUCU---ACACG-CUGCAUGCCCCCCCAACG-AGCCCCAACUUCAACUGCAGUCAUCGUCGUGUAGCAACAGUUUCCAUCC .....((((((..((..-..)).....---....(-(((((((........(((-(......(((.(....).)))..))))))))))))....)))))).... ( -23.00) >consensus CUAACGGAAACGGAGCAGAAAUCCCCU___GCUCC_GCGACUGCCUCCUGAACACAGCAACGGCUUCAACUGCAG______UCGUGUAGCAACAGUUUCCAUU_ .....(((((((((.......))).................(((..(.(((.........(((......))).........))).)..)))...)))))).... (-10.49 = -10.22 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:27 2006