
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,754,998 – 9,755,098 |
| Length | 100 |
| Max. P | 0.664746 |

| Location | 9,754,998 – 9,755,098 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 64.33 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -10.35 |
| Energy contribution | -10.77 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9754998 100 - 22224390 GCUGAAGA--GCUAUGAAAUAAGAUA-CAAACCAUUAUCAUUCGGCAGAUCCCGUUUCCUGCAGUCACUCGGCGGCUGCGAUCUUCUUCCGGCAAUCGAUGGA ((((((((--...(((..........-.....)))..)).)))))).....(((((....((((((.......))))))(((..((....))..)))))))). ( -22.26) >DroVir_CAF1 58751 83 - 1 AU------AA-----------A-CGA-UUAACCUUUG-CCUUCUAUAGAUCCCAUUGCCUGCAGUCAUUCGGCGGCUGCCAUUUUCUUUAGGCAAUCAAUGGA ..------..-----------.-...-....((.(((-(((.....(((......((...((((((.......))))))))...)))..)))))).....)). ( -16.40) >DroPse_CAF1 58835 85 - 1 AC------AA-----------GUAACAUUGACUGGUU-UUUCUUUUAGAUCCCAUUGCCUGCAGCCAUUCAGCGGCUGCCAUCUUUUUCCGACAGUCGAUGGA ..------..-----------....(((((((((...-..((.....))...........((((((.......)))))).............))))))))).. ( -23.20) >DroMoj_CAF1 65821 84 - 1 AC------AA-----------G-AUA-UAAUCCUCCGUUCCUCCGUAGAUCCCAUUGCCUGCAGUCAUUCGGCCGCGGCGAUUUUCUUCAGGCAGUCUAUGGA ..------..-----------.-...-..............(((((((((.....((((((.((....(((.(....))))....)).))))))))))))))) ( -24.40) >DroAna_CAF1 31690 90 - 1 ------------CCAAUCACUGUAACCUGAAAUGGCA-UUCUCCGCAGAUCCCAUCGCCUGCAGCCACUCUGCGGCGGCCAUCUUUUUCCGGCAGUCCAUGGA ------------(((...(((((.....((((((((.-.(((....)))......((((.((((.....))))))))))))).))).....)))))...))). ( -27.10) >DroPer_CAF1 63005 85 - 1 AC------AA-----------GAAACAUUGACUGGUU-UUUCUUUUAGAUCCCAUUGCCUGCAGCCAUUCAGCGGCUGCCAUCUUUUUCCGACAGUCGAUGGA ..------..-----------....(((((((((...-..((.....))...........((((((.......)))))).............))))))))).. ( -23.20) >consensus AC______AA___________G_AAA_UUAACCGGUA_UUUUCUGUAGAUCCCAUUGCCUGCAGCCAUUCGGCGGCUGCCAUCUUCUUCCGGCAGUCGAUGGA ...................................................((((((.((((((((.......))))))..((.......)).)).)))))). (-10.35 = -10.77 + 0.42)

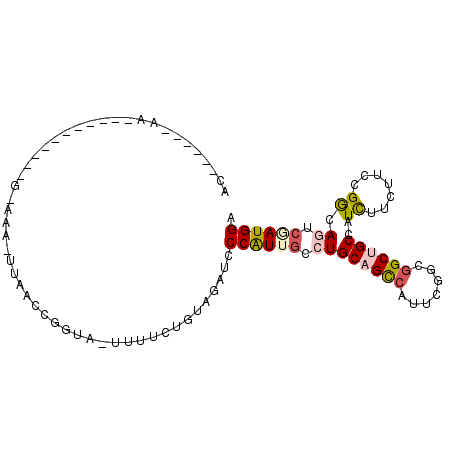
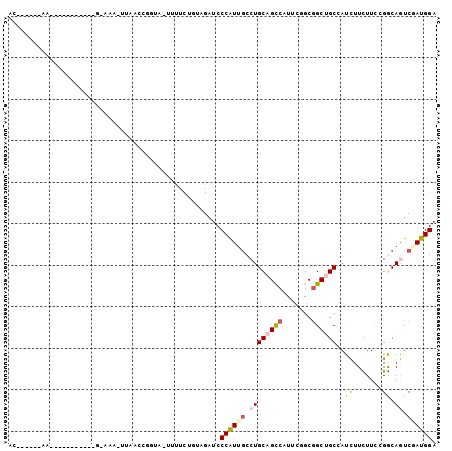
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:23 2006