| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,743,158 – 9,743,254 |
| Length | 96 |
| Max. P | 0.579366 |

| Location | 9,743,158 – 9,743,254 |
|---|---|
| Length | 96 |
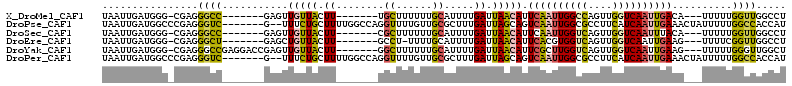
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
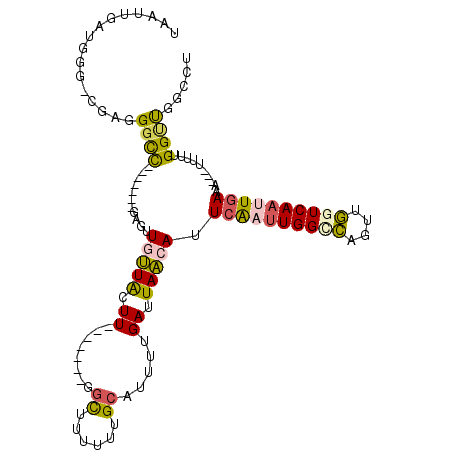
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -9.44 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9743158 96 + 22224390 UAAUUGAUGGG-CGAGGGCC-------GAGUUGUUACUU-------UGCUUUUUUGCAUUUUGAUUAACAUUCAAUUGGCCAGUUGGUCAAUUGACA---UUUUUGGUUGGCCU ...........-...(((((-------(((((((((...-------(((......)))...))).))))..((((((((((....))))))))))..---.......))))))) ( -29.00) >DroPse_CAF1 43779 105 + 1 UAAUUGAUGGCCCGAGGGUC-------G--UUUCUGCUUUUGGCCAGGUUUUGUUGCGCUUUGAUUAGCAGUCAAUUGGCGCCUUCAUCAAUUGAAACUAUUUUUGGCCACCAU .....(((((((....))))-------)--))........((((((((....(((((((((((((.....)))))..)))))..(((.....))))))....)))))))).... ( -32.00) >DroSec_CAF1 35191 96 + 1 UAAUUGAUGGG-CGAGGGCC-------GAGUUGUUACUU-------CGCUUUUUUGCAUUUUGAUUAACAUUCAAUUGGUCAGUUGGUCAAUUUACA---UUUUUGGUUGGCCU .(((((((.((-(((((((.-------((((....))))-------.)))..)))))...((((.......)))).).)))))))((((((((....---.....)))))))). ( -24.00) >DroEre_CAF1 31463 95 + 1 UAAUUGAUGGG-CGAGGGCU-------GAGCUGUUACUU-------GCCU-UUUUGCAUUUUGAUUAACAUUCACGUGGUCAGUUGGUCAAUUGAAG---UUUUCGGUUGGCCU .((((((((((-(((((((.-------.....))).)))-------))))-.....(((..(((.......))).))))))))))((((((((((..---...)))))))))). ( -28.80) >DroYak_CAF1 29629 103 + 1 UAAUUGAUGGG-CGAGGGCCGAGGACCGAGUUGUUACUU-------GGCUUUUUUGCAUUUUGAUUAACAUUCGCUUGGUCAGUUGGUCAAUUGAAG---UUUUUGGGUUGGCU ((((((((.((-((((.((.(((((((((((....))))-------)).))))).)).............))))).).))))))))(((((((.((.---...)).))))))). ( -28.74) >DroPer_CAF1 44680 105 + 1 UAAUUGAUGGCCCGAGGGUC-------G--UUUCUGCUUUUGGCCAGGUUUUGUUGCGCUUUGAUUAGCAGUCAAUUGGCGCCUUCAUCAAUUGAAACUAUUUUUGGCCACCAU .....(((((((....))))-------)--))........((((((((....(((((((((((((.....)))))..)))))..(((.....))))))....)))))))).... ( -32.00) >consensus UAAUUGAUGGG_CGAGGGCC_______GAGUUGUUACUU_______GGCUUUUUUGCAUUUUGAUUAACAUUCAAUUGGCCAGUUGGUCAAUUGAAA___UUUUUGGUUGGCCU ................((((...........(((((.((........((......)).....)).))))).((((((((((....))))))))))..........))))..... ( -9.44 = -8.80 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:22 2006