
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,743,022 – 9,743,123 |
| Length | 101 |
| Max. P | 0.979435 |

| Location | 9,743,022 – 9,743,123 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
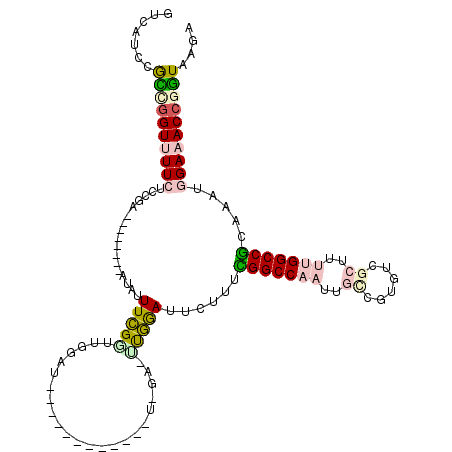
>X_DroMel_CAF1 9743022 101 + 22224390 GUCAUCCGCCGGUUUUCUCCGA--------UUAUUUCGUUUGGAU-----------UUGGAUUUGGAUUCUUUCGGCCAAUUGCCGUGACGCUUUUGGCCGCAAAUGGAAACCGGUAAGA .......(((((((((((((((--------....((((.......-----------.)))).)))))...((((((((((..((......))..))))))).))).)))))))))).... ( -35.20) >DroPse_CAF1 43636 113 + 1 GUCGUGCAUGGGUCUUGCCCAACAC--UGGAUGUUUUGGCGGUUUUCUUCAUUAAUAUUGA-CCAAAUUCUUUGGGCCAAUUG----GUCAGUGCAGGCCCUAAAUGGAAACCGGUAAGA ...(((..((((.....)))).)))--......(((((.((((((((.(((.......)))-...........(((((.(((.----...)))...))))).....)))))))).))))) ( -31.50) >DroSec_CAF1 35055 101 + 1 GUCAUCCGCCGGUUUUCUCCGA--------UUAUUUCGUUUGGAU-----------UUGGAUUUGGAUUCUUUCGGCCAAUUGCCGUGUCGCUUUUGGCCGCAAAUGGAAACCGGUAAGA .......(((((((((((((((--------....((((.......-----------.)))).)))))...((((((((((..((......))..))))))).))).)))))))))).... ( -35.20) >DroEre_CAF1 31333 95 + 1 GUCAUCCGCCCGUUUUCUCCGA--------CUGUUUCGGUUGGAU-----------------UUGGAUUCUUUCGGCCAACUGCCGUGUCGCUUUUGGCCGCAAAUGGAAACCGGUAAGA .......(((.(((((((((((--------(((...)))))))).-----------------........((((((((((..((......))..))))))).))).)))))).))).... ( -31.90) >DroYak_CAF1 29499 95 + 1 GUCAUCCGCUGGUUUUCUCCGA--------AUAUUUCGAUUGUAU-----------------UUGGAUUCUUUUGGCCAAUUGUCGUGUCGCUUUUGGCCGCAAAUGGAAACCGGUAAGU .......(((((((((((((((--------((((.......))))-----------------))))).......((((((..((......))..))))))......)))))))))).... ( -33.20) >DroPer_CAF1 44535 115 + 1 GUCGUGCAUGGGUCUUGCCCAACACUCUGGAUGUUUUGGCGGUUUUCUUCAUUAAUAUUGA-CCAAAUUCUUUGGGCCAAUUG----GUCAGUGCAGGCCCUAAAUGGAAACCGAUAAGA ...(((..((((.....)))).)))........(((((.((((((((.(((.......)))-...........(((((.(((.----...)))...))))).....)))))))).))))) ( -29.40) >consensus GUCAUCCGCCGGUUUUCUCCGA________AUAUUUCGGUUGGAU____________U_GA_UUGGAUUCUUUCGGCCAAUUGCCGUGUCGCUUUUGGCCGCAAAUGGAAACCGGUAAGA .......((((((((((.................(((((.......................)))))......(((((((..((......))..))))))).....)))))))))).... (-19.32 = -19.80 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:21 2006