
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,698,425 – 9,698,612 |
| Length | 187 |
| Max. P | 0.988480 |

| Location | 9,698,425 – 9,698,541 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9698425 116 + 22224390 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAGCAUCAACAUUGACCCAACGACAGCAACA--U-- .....(((..(((((((((((......)))))))....(((.......)))........))))...(((((((.((((.......))))..)))).))).........)))....--.-- ( -21.60) >DroPse_CAF1 38774 87 + 1 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAG--------------------------------- ...(((((...((.(((((((......))))))).)).(((.......)))........))))).......................--------------------------------- ( -15.00) >DroGri_CAF1 15884 117 + 1 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAGCAUCAACAUUGACC-AGAGACGGCAGCACAA-- .....(((.((((((((((((......)))))))............((((..........))))..(((((((.((((.......))))..)))).)))..-...))))).)))....-- ( -28.50) >DroWil_CAF1 19608 112 + 1 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAGCAUUAACAUUGCCAUAG----AGCAAGA--G-- .(((.(((...((.(((((((......))))))).)).(((.......)))........(((((.....((((.((((.......))))..)))))))))....----))).)))--.-- ( -24.60) >DroAna_CAF1 15788 118 + 1 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCCCAUGUUUUGCUACUUAUCAGCAUCAACAUUGACCGACCGACAGCAGCC--ACU .....(((.(.(((.((((.......((((((.....))))))...((((..........))))..(.(((((.((((.......))))..))))).)..)))).))).).))).--... ( -22.90) >DroPer_CAF1 42620 87 + 1 AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAG--------------------------------- ...(((((...((.(((((((......))))))).)).(((.......)))........))))).......................--------------------------------- ( -15.00) >consensus AUCUCGCUACCGUCAGUUGCACACAAUUGCAAUUAACUUGCAACACUCGCAUAAUUACAGGCGACCUCAUGUUUUGCUACUUAUCAGCAUCAACAUUGACC_A_____AGCA_CA_____ ...(((((...((.(((((((......))))))).)).(((.......)))........)))))........................................................ (-15.00 = -15.00 + 0.00)


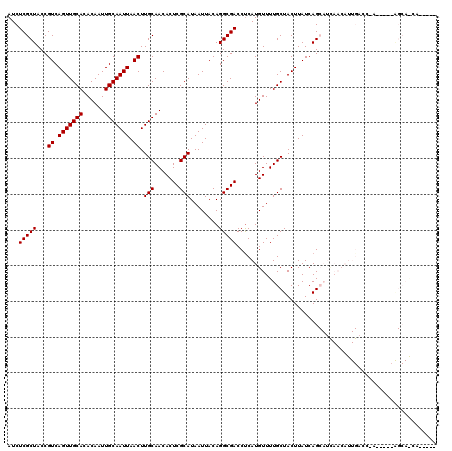
| Location | 9,698,425 – 9,698,541 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -20.33 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9698425 116 - 22224390 --A--UGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU --.--(((((((((((((((..(.(((((..(((((.....))))).)))))..)..)............(((...(((((((.....)))))))..))).)))).)))))))))).... ( -37.20) >DroPse_CAF1 38774 87 - 1 ---------------------------------CUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU ---------------------------------.......................((((((((.....((((...(((((((.....)))))))....))))....))))..))))... ( -19.30) >DroGri_CAF1 15884 117 - 1 --UUGUGCUGCCGUCUCU-GGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU --...((((((((((...-(..(.(((((..(((((.....))))).)))))..)..)...........((((...(((((((.....)))))))....))))...)))))))))).... ( -40.40) >DroWil_CAF1 19608 112 - 1 --C--UCUUGCU----CUAUGGCAAUGUUAAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU --.--(((((((----....(((.(((((..(((((.....))))).)))))...)))(((.((.....((((...(((((((.....)))))))....))))....)))))))))))). ( -33.10) >DroAna_CAF1 15788 118 - 1 AGU--GGCUGCUGUCGGUCGGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGGGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU ...--.((((((((((((((..(.(((((..(((((.....))))).)))))..)..))..........((((...(((((((.....)))))))....))))))))))))))))..... ( -44.20) >DroPer_CAF1 42620 87 - 1 ---------------------------------CUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU ---------------------------------.......................((((((((.....((((...(((((((.....)))))))....))))....))))..))))... ( -19.30) >consensus _____UG_UGCU_____U_GGUCAAUGUUGAUGCUGAUAAGUAGCAAAACAUGAGGUCGCCUGUAAUUAUGCGAGUGUUGCAAGUUAAUUGCAAUUGUGUGCAACUGACGGUAGCGAGAU ...............................(((((.....)))))..........((((((((.....((((...(((((((.....)))))))....))))....))))..))))... (-20.33 = -21.00 + 0.67)


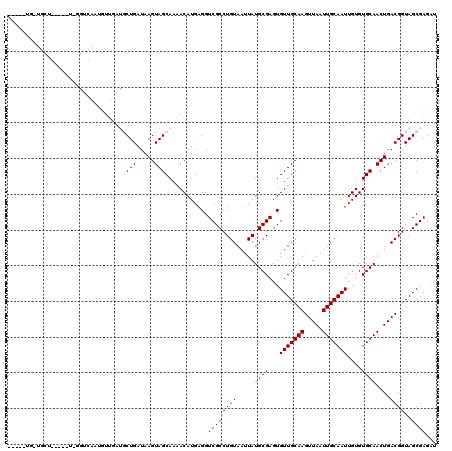
| Location | 9,698,505 – 9,698,612 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9698505 107 - 22224390 ACUGUAGCUACUCCGCUUACUA----CUACAGGGUAUCAGCGACUA-CCAUCACAAUUGCCAGCGAUUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA .((((((...............----))))))(((((((((((...-...))...((((((((((((.(((((.....))))).))))))))..)))))))))))))..... ( -37.26) >DroSec_CAF1 39996 107 - 1 ACUUUGGCUGCACCGGUUACUU----GUAUAGGGUAUCAGUGACUG-CCAUCACAAUUGCCAGCGAUAGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA ...(..((.....((((((((.----((((...)))).))))))))-..((((..((((((((((((((((((.....))))))))))))))..))))..))))))..)... ( -40.80) >DroSim_CAF1 14567 105 - 1 --UUAUGGUGCACCGGUUACUU----GUACAGGGUAUCAGUGACUG-CCAUCACAAUUGCCAGCGAUUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA --...........((((((((.----((((...)))).))))))))-.(((((..((((((((((((.(((((.....))))).))))))))..))))..)))))....... ( -36.40) >DroEre_CAF1 17702 94 - 1 -----------------UACUAUCAUGUACAGGGUAUGUAUGACUG-CCAUCACGAUGGCCGGCGAUUGCAGGUGGAUGCUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA -----------------...(((((.((((((.((.....((((((-((((....)))))(((((((.((((.......)))).)))))))))))))).))).)))))))). ( -32.30) >DroYak_CAF1 16130 108 - 1 UUAUUAGCUGCUGCGGUUACUA----GUACAGGGUAUCAUGGACUGGCCAUCGCAAUUGCCAGCGAUUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA .(((((((...((((((..(((----((.((........)).)))))..))))))....((((((((.(((((.....))))).))))))))((((....))))))))))). ( -41.40) >consensus __UUUAGCUGCACCGGUUACUA____GUACAGGGUAUCAGUGACUG_CCAUCACAAUUGCCAGCGAUUGCAGCUGGAUGUUGCUGUCGUUGGGUCAAUGUUGAUGCUGAUAA .............................((((((.......)))...(((((..((((((((((((.(((((.....))))).))))))))..))))..)))))))).... (-26.52 = -26.28 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:02 2006