
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,688,946 – 9,689,085 |
| Length | 139 |
| Max. P | 0.992964 |

| Location | 9,688,946 – 9,689,048 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.72 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9688946 102 + 22224390 GGUUUUCUCAUGCAGUUAACGCCCAGGAUCUGUCCAUAUU--UUGGAUGUCUGAGCAGGAAGC-UUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGC .(((..((.....))..)))((((((.....(((((....--.))))).(((((((((.((((-(.....((((......))))))))))))))))))))).))) ( -36.10) >DroSec_CAF1 30968 102 + 1 GGUUUUCUCAUGCAGUUAACGCCCAGGAUCUGCCCAUAUU--UUGGGUGUCUGAGCAGGAAGC-UUUUUUGUACGAAUGAGUAUAGCUUCUGCUCGGACUGUGGC .(((..((.....))..)))((((((.....(((((....--.))))).(((((((((.((((-(.....((((......))))))))))))))))))))).))) ( -38.10) >DroSim_CAF1 4981 103 + 1 GGUUUUCUCAUGCAGUUAACGCCCAGAAUCUGCCCAUAUU--UUGGGUGUCUGAGCAGGAAGCCUUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGC ...........(((((..(((((((((((........)))--))))))))((((((((.((((......(((((......))))))))))))))))))))))... ( -37.30) >DroEre_CAF1 8479 101 + 1 GCUUUUCUCAUGCAGUUAACGCCCAGGAUUUGGCCAUAUU--UUGGUUGUCUGAGCAGGAAGC-UGUUUUGUGCGAAUGAGUAUAGCUGCU-CUCGGACUGUGGC ((.........)).......((((((.....(((((....--.))))).((((((.((..(((-(((((..(....)..)).)))))).))-))))))))).))) ( -29.00) >DroYak_CAF1 6105 96 + 1 GCUUUUCUCAUGCAGUUAACGCCCAGGAUCUGCCCAUAUUUUUUGGGUGUCUG--------CC-UUUUUUGUGUGAAUGAGUAUAGCUUCUGCUCGGACUGUGGC ((..(((((((((((..((.((...((((..(((((.......))))))))))--------).-))..))))))))..(((((.......))))))))..))... ( -24.00) >consensus GGUUUUCUCAUGCAGUUAACGCCCAGGAUCUGCCCAUAUU__UUGGGUGUCUGAGCAGGAAGC_UUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGC ....................((((((.....(((((.......))))).(((((((((.((((......(((((......))))))))))))))))))))).))) (-26.60 = -27.72 + 1.12)


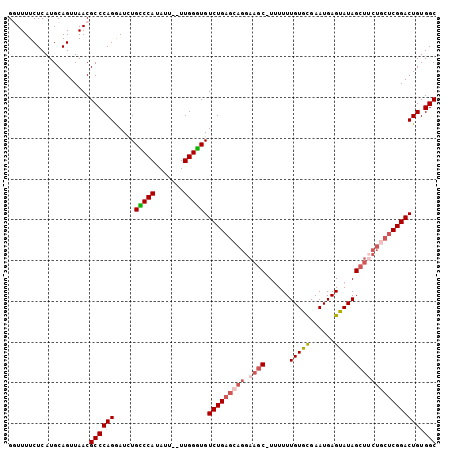
| Location | 9,688,971 – 9,689,085 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -40.34 |
| Consensus MFE | -31.76 |
| Energy contribution | -33.12 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9688971 114 + 22224390 GGAUCUGUCCAUAUU--UUGGAUGUCUGAGCAGGAAGC-UUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGCCACUUCUCAAGUUGAAAAUGAGAGGGUG---CUGCCAUAA ......(((((....--.)))))((((((((((.((((-(.....((((......)))))))))))))))))))(((((((((((((((.........))))).))))---..)))))). ( -45.20) >DroSec_CAF1 30993 114 + 1 GGAUCUGCCCAUAUU--UUGGGUGUCUGAGCAGGAAGC-UUUUUUGUACGAAUGAGUAUAGCUUCUGCUCGGACUGUGGCUACUGCUCAAGUUGAAAAUGAGAGGGUG---GUGCCACAA ......(((((....--.)))))((((((((((.((((-(.....((((......)))))))))))))))))))((((((.((..(((...............)))..---)))))))). ( -48.86) >DroSim_CAF1 5006 115 + 1 GAAUCUGCCCAUAUU--UUGGGUGUCUGAGCAGGAAGCCUUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGCCACUGCUCAAGUUGAAAAUGAGAGGGUG---GUGCCACAA ......(((((....--.)))))((((((((((.((((......(((((......)))))))))))))))))))((((((.((..(((...............)))..---)))))))). ( -47.86) >DroEre_CAF1 8504 112 + 1 GGAUUUGGCCAUAUU--UUGGUUGUCUGAGCAGGAAGC-UGUUUUGUGCGAAUGAGUAUAGCUGCU-CUCGGACUGUGGCCACUUCUCAAGAUGAAAAUGACAGGGUG----UGCUAUAU ......(((((....--.)))))(((((((.((..(((-(((((..(....)..)).)))))).))-)))))))(((((((((((.(((.........)))..)))))----.)))))). ( -33.80) >DroYak_CAF1 6130 111 + 1 GGAUCUGCCCAUAUUUUUUGGGUGUCUG--------CC-UUUUUUGUGUGAAUGAGUAUAGCUUCUGCUCGGACUGUGGCCACUUCUCAAGUUGAAAAUGAGGGGGCGGUGUUGCUAUUG ((((..(((((.......))))))))).--------..-..................(((((..((((((((.(....)))...(((((.........)))))))))))....))))).. ( -26.00) >consensus GGAUCUGCCCAUAUU__UUGGGUGUCUGAGCAGGAAGC_UUUUUUGUGCGAAUGAGUAUAGCUUCUGCUCGGACUGUGGCCACUUCUCAAGUUGAAAAUGAGAGGGUG___GUGCCAUAA ......(((((.......)))))((((((((((.((((......(((((......)))))))))))))))))))((((((..(((((((.........)))))))........)))))). (-31.76 = -33.12 + 1.36)

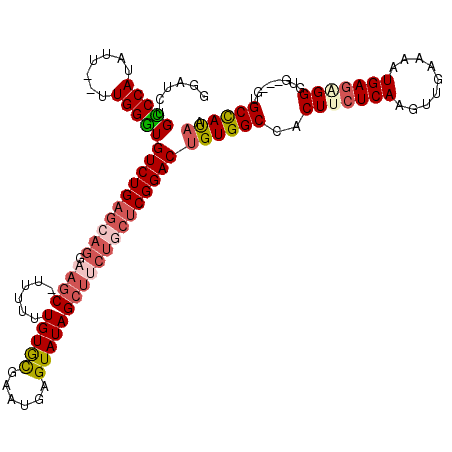
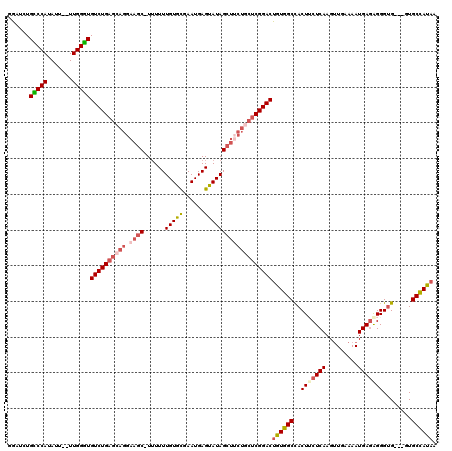
| Location | 9,688,971 – 9,689,085 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -16.37 |
| Energy contribution | -18.97 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9688971 114 - 22224390 UUAUGGCAG---CACCCUCUCAUUUUCAACUUGAGAAGUGGCCACAGUCCGAGCAGAAGCUAUACUCAUUCGCACAAAAAA-GCUUCCUGCUCAGACAUCCAA--AAUAUGGACAGAUCC ...((((..---(((..(((((.........))))).)))))))..(((.(((((((((((...................)-)))).)))))).))).((((.--....))))....... ( -33.21) >DroSec_CAF1 30993 114 - 1 UUGUGGCAC---CACCCUCUCAUUUUCAACUUGAGCAGUAGCCACAGUCCGAGCAGAAGCUAUACUCAUUCGUACAAAAAA-GCUUCCUGCUCAGACACCCAA--AAUAUGGGCAGAUCC .((((((..---.((...((((.........))))..)).))))))(((.(((((((((((.(((......)))......)-)))).)))))).))).((((.--....))))....... ( -34.40) >DroSim_CAF1 5006 115 - 1 UUGUGGCAC---CACCCUCUCAUUUUCAACUUGAGCAGUGGCCACAGUCCGAGCAGAAGCUAUACUCAUUCGCACAAAAAAGGCUUCCUGCUCAGACACCCAA--AAUAUGGGCAGAUUC .((((((..---(((...((((.........))))..)))))))))(((.(((((((((((....................))))).)))))).))).((((.--....))))....... ( -36.95) >DroEre_CAF1 8504 112 - 1 AUAUAGCA----CACCCUGUCAUUUUCAUCUUGAGAAGUGGCCACAGUCCGAG-AGCAGCUAUACUCAUUCGCACAAAACA-GCUUCCUGCUCAGACAACCAA--AAUAUGGCCAAAUCC ........----..........(((((.....))))).((((((..((.((((-((........)))..))).)).....(-((.....)))...........--....))))))..... ( -18.70) >DroYak_CAF1 6130 111 - 1 CAAUAGCAACACCGCCCCCUCAUUUUCAACUUGAGAAGUGGCCACAGUCCGAGCAGAAGCUAUACUCAUUCACACAAAAAA-GG--------CAGACACCCAAAAAAUAUGGGCAGAUCC .............(((..((((.........))))..((((....(((...(((....)))..)))...))))........-))--------).....((((.......))))....... ( -18.50) >consensus UUAUGGCAC___CACCCUCUCAUUUUCAACUUGAGAAGUGGCCACAGUCCGAGCAGAAGCUAUACUCAUUCGCACAAAAAA_GCUUCCUGCUCAGACACCCAA__AAUAUGGGCAGAUCC ...((((.....(((...((((.........))))..)))))))..(((.((((((((((......................)))).)))))).))).((((.......))))....... (-16.37 = -18.97 + 2.60)

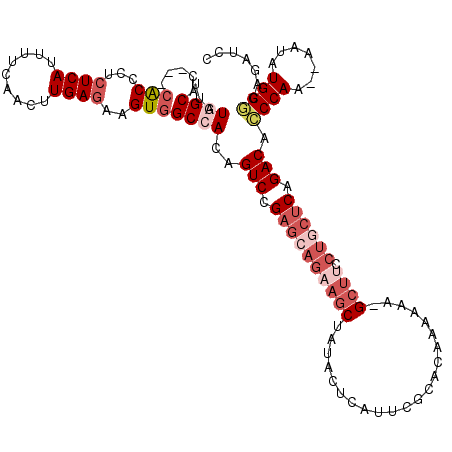

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:59 2006