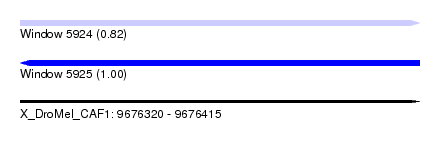
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,676,320 – 9,676,415 |
| Length | 95 |
| Max. P | 0.999221 |

| Location | 9,676,320 – 9,676,415 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9676320 95 + 22224390 UUUUGCACUUUUUAUAAAGUAUUUCAAAGGGGUGAAACAAAAAGCAGAGAACUCUGGGUUGUUUUGACCCAAAGUUUGGUUUGGCGAAAUAGUU-------A (((((((((((....)))))...(((((.(((((((((((....((((....))))..))))))).))))....)))))....)))))).....-------. ( -23.60) >DroSim_CAF1 23433 95 + 1 UUUUGCACUUUUUAUAAAGUAUUUCAAAGGGGUGAAACAAAAAGCAGAGAACUCUGGGUUGUUUUGACCCAAAGUUUGGUUUGGCGAAAUAGUU-------A (((((((((((....)))))...(((((.(((((((((((....((((....))))..))))))).))))....)))))....)))))).....-------. ( -23.60) >DroEre_CAF1 18831 94 + 1 UUUUGCGCUUUUCACAAAGUAUUUCAAAGGGGUGAAACAAAAAGCAGAGAACUCUGGGUUAUUUUGACCCAAAGUUUG-UUUGGCGAAAUAGUU-------U ((((((((((((.......(((..(...)..))).....))))))...(((((.((((((.....)))))).))))).-....)))))).....-------. ( -23.90) >DroYak_CAF1 22991 102 + 1 UUUUGCGCUUUUUAUCAAGUAUUUCAAAGGGGUGAAACAAAAAGCAGAGAAGUCUGGGUUGUUGUGACCCAAAGUUUGGUUUGGCGAAAUAGUUAAAUAGUU (((((((((((((......(((..(...)..)))....)))))))...(((.(.((((((.....)))))).).)))......))))))............. ( -21.30) >consensus UUUUGCACUUUUUAUAAAGUAUUUCAAAGGGGUGAAACAAAAAGCAGAGAACUCUGGGUUGUUUUGACCCAAAGUUUGGUUUGGCGAAAUAGUU_______A (((((((((((....))))).(((((......)))))....((((...(((((.((((((.....)))))).))))).)))).))))))............. (-20.20 = -20.70 + 0.50)

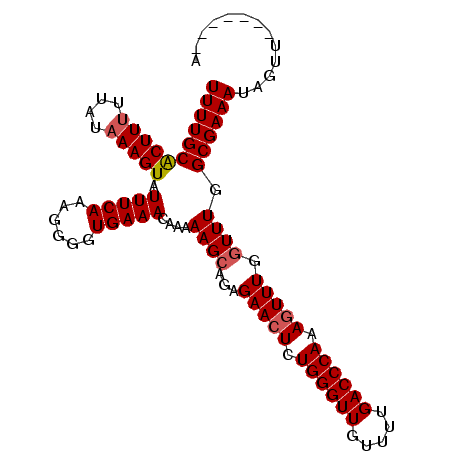
| Location | 9,676,320 – 9,676,415 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -19.77 |
| Energy contribution | -19.59 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.44 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9676320 95 - 22224390 U-------AACUAUUUCGCCAAACCAAACUUUGGGUCAAAACAACCCAGAGUUCUCUGCUUUUUGUUUCACCCCUUUGAAAUACUUUAUAAAAAGUGCAAAA .-------.........((.......((((((((((.......))))))))))....(((((((((((((......)))).......))))))))))).... ( -19.41) >DroSim_CAF1 23433 95 - 1 U-------AACUAUUUCGCCAAACCAAACUUUGGGUCAAAACAACCCAGAGUUCUCUGCUUUUUGUUUCACCCCUUUGAAAUACUUUAUAAAAAGUGCAAAA .-------.........((.......((((((((((.......))))))))))....(((((((((((((......)))).......))))))))))).... ( -19.41) >DroEre_CAF1 18831 94 - 1 A-------AACUAUUUCGCCAAA-CAAACUUUGGGUCAAAAUAACCCAGAGUUCUCUGCUUUUUGUUUCACCCCUUUGAAAUACUUUGUGAAAAGCGCAAAA .-------.........((....-..((((((((((.......))))))))))....(((((((((((((......)))))).......))))))))).... ( -22.21) >DroYak_CAF1 22991 102 - 1 AACUAUUUAACUAUUUCGCCAAACCAAACUUUGGGUCACAACAACCCAGACUUCUCUGCUUUUUGUUUCACCCCUUUGAAAUACUUGAUAAAAAGCGCAAAA .................((..........(((((((.......))))))).......(((((((((((((......))).......)))))))))))).... ( -17.31) >consensus A_______AACUAUUUCGCCAAACCAAACUUUGGGUCAAAACAACCCAGAGUUCUCUGCUUUUUGUUUCACCCCUUUGAAAUACUUUAUAAAAAGCGCAAAA .................((.......((((((((((.......))))))))))....(((((((((((((......)))))).......))))))))).... (-19.77 = -19.59 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:56 2006