| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,672,951 – 9,673,045 |
| Length | 94 |
| Max. P | 0.978105 |

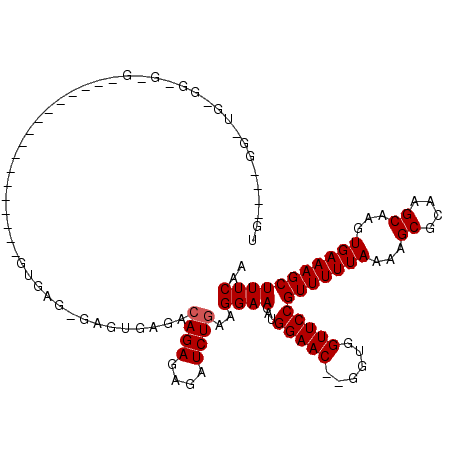
| Location | 9,672,951 – 9,673,045 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -12.17 |
| Energy contribution | -12.51 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9672951 94 + 22224390 UUGAAAGCUUUCACUUGCUUGCGCUUUUAAAAACGGAACCACC--GUUCCAUGUUCCUUCAGAUCUCUCUGUGCUACA------------------------CGCCCCGCACCCCGUCGA ((((((((........))))(((...........(((((....--))))).(((..(..((((....)))).)..)))------------------------.....)))......)))) ( -17.20) >DroEre_CAF1 15444 114 + 1 UUGAAAGCUUUCACUUGCUUGCGCUUUUAAAAACGGAACCACC--GUUCCAUGUUCCUUCAGAUCUCUCUGUCUCACUCUCUCACUCUCUCGCUCUGACCAACACUCCCCAGCC----CG ((((((((..............))))))))....(((((....--))))).((((...(((((.(..........................).)))))..))))..........----.. ( -15.01) >DroYak_CAF1 19698 90 + 1 UUGAAAGCUUUCACUUGCUUGCGCUUUUAAAAACGGAACCACCAUGUUCCAUGUUCCUUCAGAUCUCUCUCUCUCUCUCCCUCACA--------------------------AC----CA ((((((((..............))))))))....(((((......)))))....................................--------------------------..----.. ( -11.54) >consensus UUGAAAGCUUUCACUUGCUUGCGCUUUUAAAAACGGAACCACC__GUUCCAUGUUCCUUCAGAUCUCUCUGUCUCACUC_CUCAC_________________C_C_CC_CA_CC____CA ((((((((..............))))))))....(((((......))))).........((((....))))................................................. (-12.17 = -12.51 + 0.33)

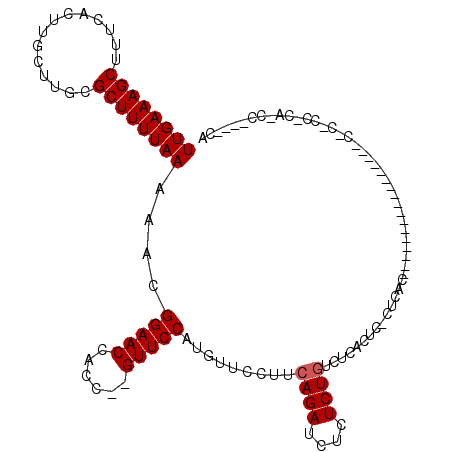
| Location | 9,672,951 – 9,673,045 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -18.73 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9672951 94 - 22224390 UCGACGGGGUGCGGGGCG------------------------UGUAGCACAGAGAGAUCUGAAGGAACAUGGAAC--GGUGGUUCCGUUUUUAAAAGCGCAAGCAAGUGAAAGCUUUCAA .....(..((((...(((------------------------(......((((....))))...(((.(((((((--....))))))).)))....))))..))).((....)))..).. ( -24.70) >DroEre_CAF1 15444 114 - 1 CG----GGCUGGGGAGUGUUGGUCAGAGCGAGAGAGUGAGAGAGUGAGACAGAGAGAUCUGAAGGAACAUGGAAC--GGUGGUUCCGUUUUUAAAAGCGCAAGCAAGUGAAAGCUUUCAA ..----(((..(......)..)))...............((((((....((((....))))...(((.(((((((--....))))))).)))....((....))........)))))).. ( -27.70) >DroYak_CAF1 19698 90 - 1 UG----GU--------------------------UGUGAGGGAGAGAGAGAGAGAGAUCUGAAGGAACAUGGAACAUGGUGGUUCCGUUUUUAAAAGCGCAAGCAAGUGAAAGCUUUCAA ..----..--------------------------...............(((((...((.....(((.(((((((......))))))).)))....((....))....))...))))).. ( -19.30) >consensus UG____GG_UG_GG_G_G_________________GUGAG_GAGUGAGACAGAGAGAUCUGAAGGAACAUGGAAC__GGUGGUUCCGUUUUUAAAAGCGCAAGCAAGUGAAAGCUUUCAA .................................................((((....))))..((((...(((((......)))))(((((((...((....))...))))))))))).. (-18.73 = -19.07 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:50 2006