
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,669,164 – 9,669,353 |
| Length | 189 |
| Max. P | 0.892228 |

| Location | 9,669,164 – 9,669,283 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -28.64 |
| Energy contribution | -29.83 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9669164 119 - 22224390 GUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGCCAUUUAAAAUGGGCAGUGAGCGUGUUGCAGCUUCAAAUUUUCACAGGGUAACUGAAAUCGCUAGUUUUCACGAUUUUGUC (.((((((.(((..(..(((((((.(((..(((....)))....))).)))))))..)..).)).)))))).)........(((((((...(((((.(....).)))))..))))))). ( -34.90) >DroSim_CAF1 18325 119 - 1 GUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGCCAUUUAAAAUGGGCAGUGAGCGUGUUGCAGCUUCAAAUUUUCACAGGGUAACUGAAAUCGGUAGAUCCUAUGAUUUUGUC ..((((((.(((..(..(((((((.(((..(((....)))....))).)))))))..)..).)).)))))).((((...(((.(((((.((((....))))..))))).))).)))).. ( -39.70) >DroEre_CAF1 11661 119 - 1 GUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGCCAUUUAAAAUGGGCAGUGAGCGUGUUGCGGCUUCAAAUUCGUGCAGGGUAACUGAAAUCGGUACUUUCUACGGUUUCUUC (.((((((.(((..(..(((((((.(((..(((....)))....))).)))))))..)..).)).)))))).)....(((((.((((((.(((....))))))))).)))))....... ( -38.50) >DroYak_CAF1 15861 100 - 1 GUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGCCAUUUAAAAUGGGCAGUGAGCGUGUUGCAGCUUCACAUUUUUACAGGGUACGCAAAUUCG-U------------------ ((((((((.(((..(..(((((((.(((..(((....)))....))).)))))))..)..).)).)))))..))).........((((......)))).-.------------------ ( -29.30) >consensus GUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGCCAUUUAAAAUGGGCAGUGAGCGUGUUGCAGCUUCAAAUUUUCACAGGGUAACUGAAAUCGGUAGUUUCUACGAUUUUGUC (.((((((.(((..(..(((((((.(((..(((....)))....))).)))))))..)..).)).)))))).)...................(((((((..........)))))))... (-28.64 = -29.83 + 1.19)



| Location | 9,669,244 – 9,669,353 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9669244 109 + 22224390 GCAACAGCCGAAAAGUGAACAGCAAUUGGCAGCAGCCACCAAUUCAGGAGAGGCUUCACAAAAUGGAAACAGAAGUGCUGUGAGAGG---------GGGGGUAGAGAUGCUGGUGGAG .......((.....(....)....((..(((.(.(((.((..(((.(.((..(((((......((....))))))).)).)..))).---------.))))).)...)))..)))).. ( -25.80) >DroSim_CAF1 18405 116 + 1 GCAACAGCCGAAAAGUGAACAGCAAUUGGCAGCAGCCACCAAUUCAGGAGAGGCUUCACAAAAUGGAAACAGAAGUGCUGUGAAGGGGAGGGG--GAGGGGUGGAGAAGCUGGCGGAG ((....(((((...((.....))..)))))(((..(((((..(((........(((((((.(.((....))....)..))))))).......)--))..)))))....))).)).... ( -32.96) >DroEre_CAF1 11741 113 + 1 GCAACAGCCGAAAAGUGAACAGCAAUUGGCAGCAGCCACCAAUUCAGGAGAGGCUUCACAAAAUGGAAACAGAAGUGCUGUGAAGGG-----GUUGGGGGUUGGAGAGGCUGGUGGAG .((.(((((............((.....))..(((((.(((((((........(((((((.(.((....))....)..)))))))))-----))))).)))))....))))).))... ( -35.50) >DroYak_CAF1 15922 118 + 1 GCAACAGCCGAAAAGUGAACAGCAAUUGGCAGCAGCCACCAAUUCAGGAGAGCCUUCACAAAAUGGACACAGAAGUGCUGUGAAGGGUGGGGGUGUGGGUGUGGAGAAGCUGGUGGAG ......(((((...((.....))..))))).....((((((.((((.(...((((((((...((((.(((....))))))).....)))))))).....).)))).....)))))).. ( -34.20) >consensus GCAACAGCCGAAAAGUGAACAGCAAUUGGCAGCAGCCACCAAUUCAGGAGAGGCUUCACAAAAUGGAAACAGAAGUGCUGUGAAGGG_____G__GGGGGGUGGAGAAGCUGGUGGAG ......(((((...((.....))..))))).....((((((.(((....))).(((((((.(.((....))....)..))))))).........................)))))).. (-22.91 = -23.23 + 0.31)

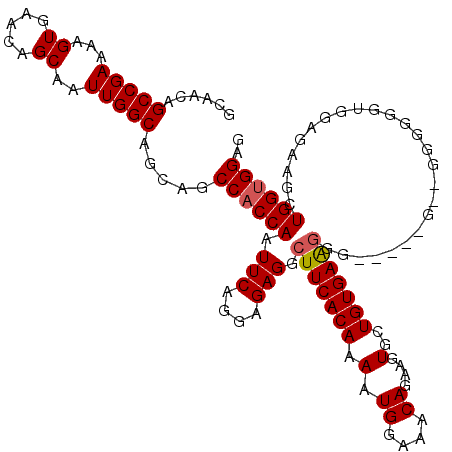
| Location | 9,669,244 – 9,669,353 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9669244 109 - 22224390 CUCCACCAGCAUCUCUACCCCC---------CCUCUCACAGCACUUCUGUUUCCAUUUUGUGAAGCCUCUCCUGAAUUGGUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGC ......((((............---------....((((((................))))))((((...((......)).)))))))).((((.((((.........)))).)))). ( -22.29) >DroSim_CAF1 18405 116 - 1 CUCCGCCAGCUUCUCCACCCCUC--CCCCUCCCCUUCACAGCACUUCUGUUUCCAUUUUGUGAAGCCUCUCCUGAAUUGGUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGC ......((((....(((((..((--........((((((((................))))))))........))...)))))..)))).((((.((((.........)))).)))). ( -24.88) >DroEre_CAF1 11741 113 - 1 CUCCACCAGCCUCUCCAACCCCCAAC-----CCCUUCACAGCACUUCUGUUUCCAUUUUGUGAAGCCUCUCCUGAAUUGGUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGC ...((.(((((...((((........-----..((((((((................))))))))..((....)).)))).))))).)).((((.((((.........)))).)))). ( -24.79) >DroYak_CAF1 15922 118 - 1 CUCCACCAGCUUCUCCACACCCACACCCCCACCCUUCACAGCACUUCUGUGUCCAUUUUGUGAAGGCUCUCCUGAAUUGGUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGC ......((((....((((..............((((((((((((....))))......)))))))).............))))..)))).((((.((((.........)))).)))). ( -29.53) >consensus CUCCACCAGCUUCUCCACCCCCC__C_____CCCUUCACAGCACUUCUGUUUCCAUUUUGUGAAGCCUCUCCUGAAUUGGUGGCUGCUGCCAAUUGCUGUUCACUUUUCGGCUGUUGC ...((.(((((......................((((((((................)))))))).......(((((...((((....))))......)))))......))))).)). (-20.82 = -20.82 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:46 2006