
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,668,295 – 9,668,454 |
| Length | 159 |
| Max. P | 0.971610 |

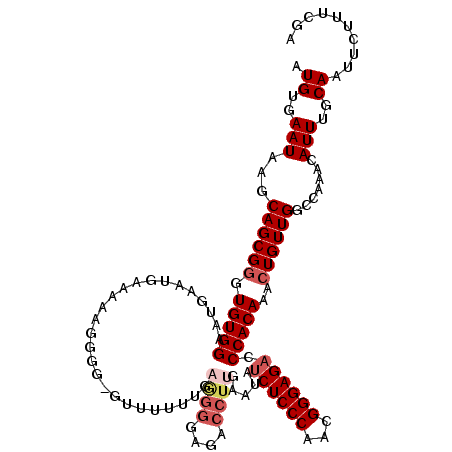
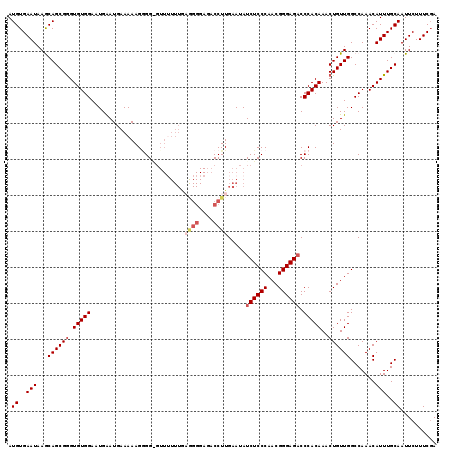
| Location | 9,668,295 – 9,668,414 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -25.30 |
| Energy contribution | -26.58 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9668295 119 - 22224390 AUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGUGGGGUUUUUUAAGGGGAGACCUUGAAUAUCUCCCAACGGGAGUCCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUCGA ...((((..((..((((((((((.............((((((...(((((((.....)))))))...(((((...)))))))))))...(....).))..))))))))..))..)))). ( -31.90) >DroSec_CAF1 3906 119 - 1 AUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGGGGAGUUUUUUGAGGGGAGACCUUGAAUAUCUCCCAACGGGAGACCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUCGA .((..(((..(((((((.((((...............(((((....((.((((....)))).))...)))))...((....))))))..))))).)).....)))..)).......... ( -35.50) >DroSim_CAF1 17490 117 - 1 AUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGAGGGGUU-UUUGAAGGGAGACCUUGAAUAUCUCCCAACGGGAGACCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUC-A .((..(((..(((((((.(((((.....................-.((.((((....)))).))..((((((...)))))).)))))..))))).)).....)))..))........-. ( -32.30) >DroEre_CAF1 10792 118 - 1 AUGUGAAUAAACAGCGAGUGUGGAAUGGAUGAAAAAGGGG-GUUUUUUGCGGGGAGACCUUGAAUAUCUCCCAACGGGAGACCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUCGA ...((((..((..((((((((....(((..((...((.((-((((((((..((((((.........))))))..)))))))))).....)).))..))).))))))))..))..)))). ( -38.30) >DroYak_CAF1 15022 104 - 1 AUGUGAAUAAACAGCGGGUGUGGAAUGAAUGAAAAAGGGG---------------AACCUUGAAUAUCUCCCAACGGGAGACCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUCGA .((..(((...((((((.(((((...........((((..---------------..)))).....((((((...)))))).)))))..)))))).......)))..)).......... ( -30.40) >consensus AUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGGGG_GUUUUUUGAGGGGAGACCUUGAAUAUCUCCCAACGGGAGACCCACAAACUGUUGGCCAAACAUUUGCAAUUCUUUCGA .((..(((...((((((.(((((..........................((((....)))).....((((((...)))))).)))))..)))))).......)))..)).......... (-25.30 = -26.58 + 1.28)

| Location | 9,668,334 – 9,668,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.13 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9668334 120 + 22224390 CUCCCGUUGGGAGAUAUUCAAGGUCUCCCCUUAAAAAACCCCACUUUUUCAUUCAUUCCACACCCGCUGCUUAUUCACAUUCGUUUUUAUUUUUUGUUACUGAAUGUGGGGGAGAUGAAA (((((...(((((((.......))))))).............................................(((((((((.(..((.....))..).))))))))))))))...... ( -27.90) >DroSec_CAF1 3945 118 + 1 CUCCCGUUGGGAGAUAUUCAAGGUCUCCCCUCAAAAAACUCCCCUUUUUCAUUCAUUCCACACCCGCUGCUUAUUCACAUUCGUUUUUAUUUUUUGUUACUGGGUGUGGGGG--AUGAAA .....(..(((((((.......)))))))..)..............((((((((...(((((((((..((.........................))...))))))))).))--)))))) ( -33.61) >DroSim_CAF1 17528 117 + 1 CUCCCGUUGGGAGAUAUUCAAGGUCUCCCUUCAAA-AACCCCUCUUUUUCAUUCAUUCCACACCCGCUGCUUAUUCACAUUCGUUUUUAUUUUUUGUUACUGGGUGUGGGGG--AUGAAA .....(..(((((((.......)))))))..)...-..........((((((((...(((((((((..((.........................))...))))))))).))--)))))) ( -33.21) >DroEre_CAF1 10831 117 + 1 CUCCCGUUGGGAGAUAUUCAAGGUCUCCCCGCAAAAAAC-CCCCUUUUUCAUCCAUUCCACACUCGCUGUUUAUUCACAUUCGUUUUUAUUUUUCGCUUCUGGGUGUGAGGA--AUGAAA .....((.(((((((.......))))))).)).......-.............(((((((((.....)))....(((((((((.................))))))))))))--)))... ( -27.63) >DroYak_CAF1 15061 103 + 1 CUCCCGUUGGGAGAUAUUCAAGGUU---------------CCCCUUUUUCAUUCAUUCCACACCCGCUGUUUAUUCACAUUCGGUUUUAUUUUUGGUUUCUGGGUGUGAGGG--AUGAAU (((((...)))))......((((..---------------..))))....((((((((((((.....)))....((((((((((..(((....)))...)))))))))))))--)))))) ( -29.30) >consensus CUCCCGUUGGGAGAUAUUCAAGGUCUCCCCUCAAAAAAC_CCCCUUUUUCAUUCAUUCCACACCCGCUGCUUAUUCACAUUCGUUUUUAUUUUUUGUUACUGGGUGUGGGGG__AUGAAA ........(((((((.......)))))))..................(((((...(((((((((((..((.........................))...)))))))))))...))))). (-22.21 = -23.13 + 0.92)



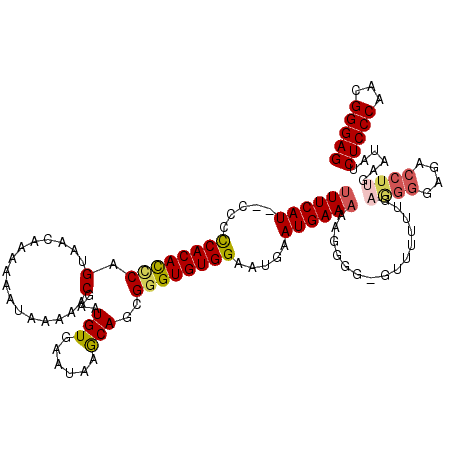
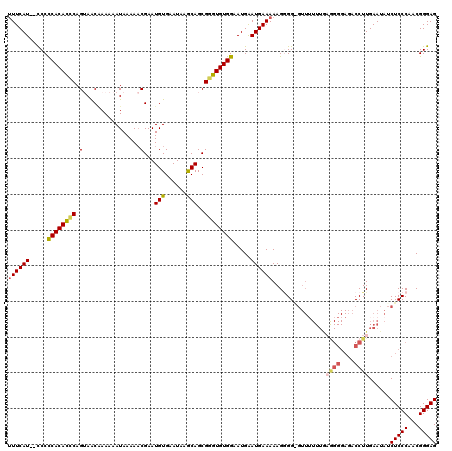
| Location | 9,668,334 – 9,668,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.59 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9668334 120 - 22224390 UUUCAUCUCCCCCACAUUCAGUAACAAAAAAUAAAAACGAAUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGUGGGGUUUUUUAAGGGGAGACCUUGAAUAUCUCCCAACGGGAG ......(((((((((((((.((..............))...(((......)))..))))))))..............(((((...(((((((.....)))))))...).))))..))))) ( -31.64) >DroSec_CAF1 3945 118 - 1 UUUCAU--CCCCCACACCCAGUAACAAAAAAUAAAAACGAAUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGGGGAGUUUUUUGAGGGGAGACCUUGAAUAUCUCCCAACGGGAG .....(--(((((((((((.((..............))...(((......)))..))))))))..............(((((....((.((((....)))).))...)))))...)))). ( -39.64) >DroSim_CAF1 17528 117 - 1 UUUCAU--CCCCCACACCCAGUAACAAAAAAUAAAAACGAAUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGAGGGGUU-UUUGAAGGGAGACCUUGAAUAUCUCCCAACGGGAG .....(--(((((((((((.((..............))...(((......)))..)))))))).............(.((((..-.((.((((....)))).))...)))))...)))). ( -36.14) >DroEre_CAF1 10831 117 - 1 UUUCAU--UCCUCACACCCAGAAGCGAAAAAUAAAAACGAAUGUGAAUAAACAGCGAGUGUGGAAUGGAUGAAAAAGGGG-GUUUUUUGCGGGGAGACCUUGAAUAUCUCCCAACGGGAG .(((((--(((.(((...(......)...........((..(((......))).)).))).)))))))).......((((-(...((((.(((....)))))))...)))))........ ( -28.70) >DroYak_CAF1 15061 103 - 1 AUUCAU--CCCUCACACCCAGAAACCAAAAAUAAAACCGAAUGUGAAUAAACAGCGGGUGUGGAAUGAAUGAAAAAGGGG---------------AACCUUGAAUAUCUCCCAACGGGAG ((((((--...((((((((.(....)...............(((......)))..)))))))).))))))....((((..---------------..))))......(((((...))))) ( -27.10) >consensus UUUCAU__CCCCCACACCCAGUAACAAAAAAUAAAAACGAAUGUGAAUAAGCAGCGGGUGUGGAAUGAAUGAAAAAGGGG_GUUUUUUGAGGGGAGACCUUGAAUAUCUCCCAACGGGAG ((((((.....((((((((.(................)...(((......)))..)))))))).....))))))...............((((....))))......(((((...))))) (-23.07 = -23.59 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:41 2006