
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,665,538 – 9,665,631 |
| Length | 93 |
| Max. P | 0.804552 |

| Location | 9,665,538 – 9,665,631 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
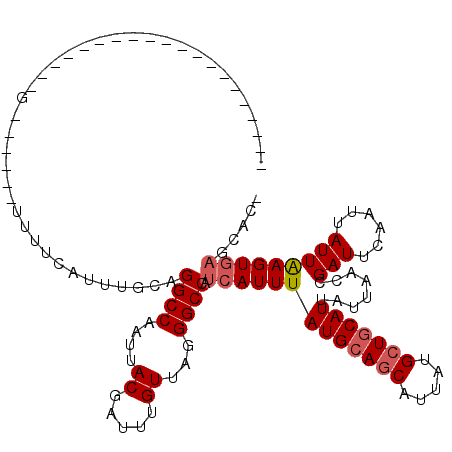
>X_DroMel_CAF1 9665538 93 + 22224390 --------------------G------UUUUCAUUUGCAGGCCAAUUACGAUUUGUUGGGGCCAUCAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAGCUC- --------------------.------..((((((((..((((....((.....))...)))).......(((((((.....)))))))...................))))))))...- ( -23.00) >DroSec_CAF1 1203 93 + 1 --------------------G------UUUUCAUUUGCAGGCCAAUUACGAUUUGUUAGGGCCAUCAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAGCAC- --------------------.------........(((.((((....((.....))...)))).(((((((((((((.....)))))))........(((......)))))))))))).- ( -23.30) >DroAna_CAF1 29511 120 + 1 CGUUCUCAGUUUAAUUUUCGGUUUCCAUUUUCGCUUUCGGGCCAAUUACGUUUUGUUAGGGCCAUAAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUGAGUGCCCCCC .((.((((((.......(((((.................((((....((.....))...)))).((((..(((((((.....))))))).))))))))).......)))))).))..... ( -33.24) >DroSim_CAF1 15607 93 + 1 --------------------G------UUUUCAUUUGCAGGCCAAUUACGAUUUGUUAGGGCCAUCAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAGCAC- --------------------.------........(((.((((....((.....))...)))).(((((((((((((.....)))))))........(((......)))))))))))).- ( -23.30) >DroEre_CAF1 8020 96 + 1 -----------------UUGG------UUUCCAUUUGCAGGCCAAUUACGAUUUGUUAGGGCCAUCAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAACAG- -----------------((((------((..........((((....((.....))...)))).......(((((((.....)))))))....))))))....................- ( -22.60) >DroYak_CAF1 12261 83 + 1 ------------------------------------GCUGGCCAAUUACGAUUUGUUAGGGCCAUCAUUUAUGAAACAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAGCAG- ------------------------------------(((((((....((.....))...)))))(((((((((((.(....................).)))).....)))))))))..- ( -13.15) >consensus ____________________G______UUUUCAUUUGCAGGCCAAUUACGAUUUGUUAGGGCCAUCAUUUAUGCAGCAUUAUGCUGCAUUAUUAACCGAUUCAAUUAUUAAGUGAGCAC_ .......................................((((....((.....))...)))).(((((((((((((.....)))))))........(((......)))))))))..... (-17.00 = -17.53 + 0.53)

| Location | 9,665,538 – 9,665,631 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
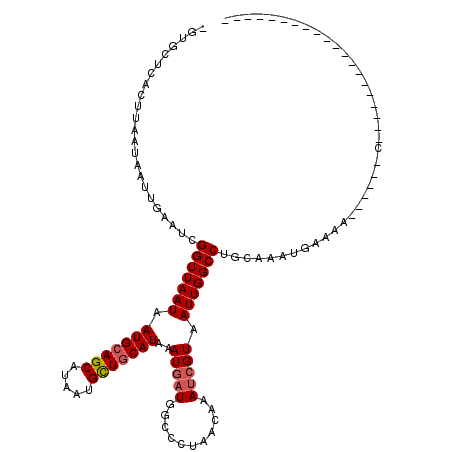
>X_DroMel_CAF1 9665538 93 - 22224390 -GAGCUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUGAUGGCCCCAACAAAUCGUAAUUGGCCUGCAAAUGAAAA------C-------------------- -..(((((.........)))...(((((((.(((((((.....)))))))..(((((...........))))).))))))).)).........------.-------------------- ( -19.80) >DroSec_CAF1 1203 93 - 1 -GUGCUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUGAUGGCCCUAACAAAUCGUAAUUGGCCUGCAAAUGAAAA------C-------------------- -.((((((.........)))...(((((((.(((((((.....)))))))..(((((...........))))).))))))).)))........------.-------------------- ( -20.50) >DroAna_CAF1 29511 120 - 1 GGGGGGCACUCAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUUAUGGCCCUAACAAAACGUAAUUGGCCCGAAAGCGAAAAUGGAAACCGAAAAUUAAACUGAGAACG ........((((.(((((...((((((....(((((((.....))))))).......((((...((.....))....))))................)))))).)))))...)))).... ( -28.80) >DroSim_CAF1 15607 93 - 1 -GUGCUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUGAUGGCCCUAACAAAUCGUAAUUGGCCUGCAAAUGAAAA------C-------------------- -.((((((.........)))...(((((((.(((((((.....)))))))..(((((...........))))).))))))).)))........------.-------------------- ( -20.50) >DroEre_CAF1 8020 96 - 1 -CUGUUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUGAUGGCCCUAACAAAUCGUAAUUGGCCUGCAAAUGGAAA------CCAA----------------- -..(((((.........))))).((((....(((((((.....)))))))...((..((((...((.....))....))))..))......))------))..----------------- ( -21.60) >DroYak_CAF1 12261 83 - 1 -CUGCUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGUUUCAUAAAUGAUGGCCCUAACAAAUCGUAAUUGGCCAGC------------------------------------ -.((((((.(((.((((((...)))))).))))).)))).................(((((...((.....))....)))))..------------------------------------ ( -13.50) >consensus _GUGCUCACUUAAUAAUUGAAUCGGUUAAUAAUGCAGCAUAAUGCUGCAUAAAUGAUGGCCCUAACAAAUCGUAAUUGGCCUGCAAAUGAAAA______C____________________ .......................(((((((.(((((((.....)))))))..(((((...........))))).)))))))....................................... (-16.72 = -17.08 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:38 2006