
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,660,313 – 9,660,538 |
| Length | 225 |
| Max. P | 0.856761 |

| Location | 9,660,313 – 9,660,426 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.05 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -24.72 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
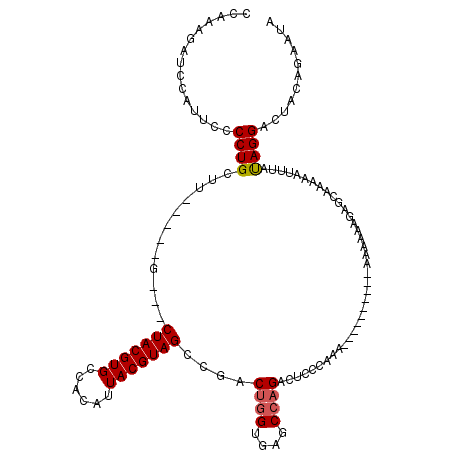
>X_DroMel_CAF1 9660313 113 - 22224390 AGCCAAAAAAGGCCAGGUAAGCUGCACUAAAAGAAGCAACUAUUUCGUGUUCCUUAUAUUUAUAGUCAAGCGACAAUAAGUUAUAGUAAGCAAAGGCGGCAUUGAGUCAUAAC .(((......))).......(((((.((....((((......)))).((((..((((((((((.(((....))).))))).)))))..)))).)))))))............. ( -22.90) >DroSim_CAF1 10359 113 - 1 AGCCAAAAAAGACCAGGUAAGCUGCACUAAAAGAAGCAACUAUUUCGUGUUGCUUAUAUUUAUAGUCAAGCGACAAUAAGUUAUAGUAAGCAAAGGCGGCAUUGAGUCAUAAC ..........((((((....(((((.((....((((......))))...((((((((((((((.(((....))).))))))....))))))))))))))).))).)))..... ( -26.80) >DroEre_CAF1 2836 113 - 1 AGCCAAAAAAGACCAGGUAAGCUGCACUAAAAGAAGCAACUAUUCUGUGUUGCUUAUAUUUAUAGUCGAGCGACAAUAAGUUAUAGUAAGCAAAGGCGGCAUUGAGUCAUAAC ..........((((((....(((((.((...((((.......))))...((((((((((((((.(((....))).))))))....))))))))))))))).))).)))..... ( -27.90) >consensus AGCCAAAAAAGACCAGGUAAGCUGCACUAAAAGAAGCAACUAUUUCGUGUUGCUUAUAUUUAUAGUCAAGCGACAAUAAGUUAUAGUAAGCAAAGGCGGCAUUGAGUCAUAAC ..........((((((....(((((.((.....(((((((........)))))))..((((((.(((....))).))))))............))))))).))).)))..... (-24.72 = -24.83 + 0.11)



| Location | 9,660,426 – 9,660,538 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.62 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -13.81 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9660426 112 - 22224390 CCAAAGAUCCAUUCCCCUGCUC-----G---CUACGUGCCACAUUACGUAGCCGACUGGUGAGCCAGACUCCCAAAAAAAAGGAAAAAAAAAGAGCAAAAAUUUAUAGGACUACAGAAUA ....((.(((((.....(((((-----(---(((((((......))))))))...((((....))))..(((.........)))........))))).......)).)))))........ ( -25.50) >DroAna_CAF1 24827 97 - 1 ----UCCUGCACUCUCCUGUUUCGUUAG---CUACGUGCCACAUUACGUAGCCGACCUGUGCGCCAGACA---AAA----------AACAAAGGGCGAAGCCAAGCAGGAGCAG---CUA ----....((...(((((((((.((..(---(((((((......))))))))..))((...((((.....---...----------.......)))).))..)))))))))..)---).. ( -32.36) >DroSim_CAF1 10472 105 - 1 CCAAAGAUCCAUACCCCUGCUU-----GCUGCUACGUGCCACAUUACGUAGCCGACUGGUGAGCCAGACUCCAAAA----------AAAAAAGAGCAAAAAUUUAUAGGACUACAGAAUA ....((.((((((....(((((-----.(.((((((((......)))))))).).((((....)))).........----------......)))))......))).)))))........ ( -23.30) >DroEre_CAF1 2949 99 - 1 CCGAAGAUCCAUUCCCCUGCCU-----G---CUACGUGCCACAUUACGUAGCCGACUGGUAAGCCAGACUCCCAAA-------------AAAGAGCACAAAUUUAUAGGACUAGAGAGUA ..........((((.(..((((-----(---(((((((......))))))))...((((....)))).(((.....-------------...)))...........))).)..).)))). ( -21.40) >consensus CCAAAGAUCCAUUCCCCUGCUU_____G___CUACGUGCCACAUUACGUAGCCGACUGGUGAGCCAGACUCCCAAA__________AAAAAAGAGCAAAAAUUUAUAGGACUACAGAAUA ...............((((............(((((((......)))))))....((((....))))......................................))))........... (-13.81 = -14.12 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:34 2006